Intracellular Developmental Programs Can Help Determine the Time-Course of a Cell’s Development
Although signaling between cells plays an essential part in driving the progress of development, this does not mean that cells always need signals from other cells to prod them into changing their character as development proceeds. Some of these changes are intrinsic to the cell (like the ticking of the segmentation clock) and depend on intracellular developmental programs that can operate even when the cell is removed from its normal environment. The best-understood example is in the development of neural precursor cells, or neuroblasts, in the embryonic Drosophila central nervous system. These cells, as we saw, are initially singled out from the neurogenic ectoderm of the embryo by a typical lateral-inhibition mechanism that depends on Notch, and they then proceed through an entirely predictable series of asymmetric cell divisions to generate ganglion mother cells that divide to form neurons and glial cells.
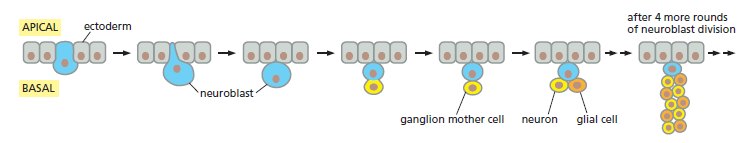
Neuroblasts and asymmetric cell division in the central nervous system of a fly embryo. The neuroblast originates as a specialized ectodermal cell. It is singled out by lateral inhibition and emerges from the basal (internal) face of the ectoderm. It then goes through repeated division cycles, dividing asymmetrically to generate a series of ganglion mother cells. Each ganglion mother cell divides just once to give a pair of differentiated daughters (typically a neuron plus a glial cell).
The neuroblast changes its internal state as it goes through its set program of divisions, generating different cell types with a reproducible sequence and timing. These successive changes in neuroblast specification occur through the sequential expression of specific transcription regulators. For example, most embryonic neuroblasts sequentially express the transcription regulators Hunchback, Krüppel, Pdm, and Cas in a fixed order

Temporal patterning of neuroblast fate in Drosophila. Hunchback, Krüppel, Pdm, and Cas are transcription regulators that are expressed consecutively in the cell lineage of neuroblasts during development of the Drosophila nervous system. At successive time steps, correlated with cell division, the neuroblast switches its pattern of gene expression. Each neuroblast division produces one daughter that remains a neuroblast and expresses the updated set of genes, and one ganglion mother cell that maintains the expression of this gene set and differentiates into specific cell types accordingly.
When a neuroblast divides, the set of transcription regulators expressed at that time is inherited by the ganglion mother cell and its neural progeny; thus, the differentiated neural cells are endowed with different characters according to their time of birth. Remarkably, when neuroblasts are taken from an embryo and maintained in culture, isolated from their normal surroundings, they step through much the same stereotyped developmental program as if they had been left in the embryo. Moreover, many of the neuroblast transitions occur even when cell division is blocked. The neuroblasts seem to have a built-in timer how was this timer programmed ? that determines when each of the transcription regulators is expressed, and this timer can continue to run in the absence of cell division. The molecular basis of the timing is largely unknown;
in part, at least, it must depend on the time taken for gene switching, as described above; but it may well also depend on slow progressive changes in chromatin structure. These too can serve to measure the passage of time in the embryo.
Cells Rarely Count Cell Divisions to Time Their Development
Many specialized cells in animals develop from proliferating progenitor cells that stop dividing and terminally differentiate after a limited number of cell divisions. In these cases, differentiation is coordinated how was this coordination programmed ? with withdrawal from the cell cycle, but it is usually not known how the coordination is achieved. It has often been suggested that the cell-division cycle might serve as an intracellular timer to control the timing of cell differentiation. The cell cycle would be the ticking clock that sets the tempo of other developmental processes, with maturational changes in gene expression being dependent on cell-cycle progression. Most of the evidence, however, indicates that this tempting idea is wrong. Although there are examples where cells change their maturation state with each division and the change depends on cell division, this is not the general rule. As we just saw for neuroblasts in the Drosophila embryo, cells in developing animals often carry on with their normal timetable of maturation and differentiation even when cell division is artificially blocked; necessarily, some abnormalities occur, if only because a single undivided cell cannot differentiate in two ways at once. But it seems that most developing cells can change their state without a requirement for cell division. Developmental control genes can switch the cell-division-cycle machinery on or off, and it is the dynamics of these genes, rather than the cell cycle, that sets the tempo of development.
MicroRNAs Often Regulate Developmental Transitions
Genetic screens are useful for tracking down the genes involved in almost any biological process, and they have been used to search for mutations that alter developmental
timing. Such screens were performed in the nematode Caenorhabditis elegans
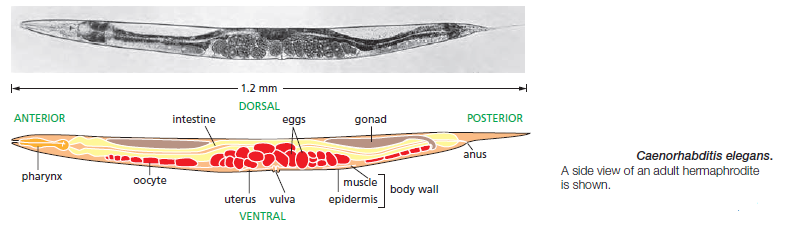
This worm is small, relatively simple, and precisely structured. The anatomy of its development is highly predictable and has been described in extraordinary detail, so that one can map out the exact lineage of every cell in the body and see exactly how the developmental program is altered in a mutant. Genetic screens in C. elegans revealed mutations that disrupt developmental timing in a particularly striking way: in these so-called heterochronic mutants, certain cells in a larva at one stage of development behave as though they were in a larva at a different stage of development, or cells in the adult carry on dividing as though they belonged to a larva
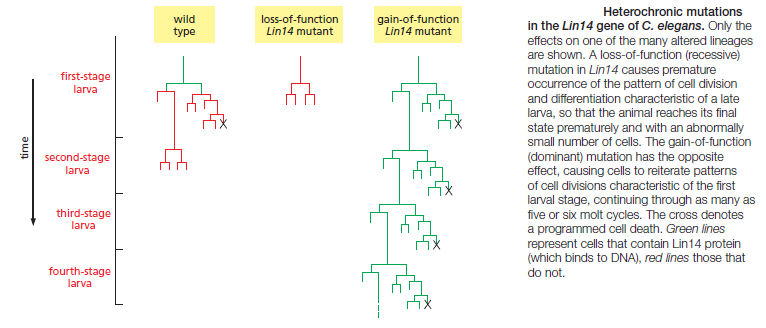
Genetic analyses showed that the products of the heterochronic genes act in series, forming regulatory cascades. Unexpectedly, two genes at the top of their respective cascades, called Lin4 and Let7, were found to code not for protein but instead for microRNAs (miRNAs)—short, untranslated, regulatory RNA molecules, 21 or 22 nucleotides long. These act by binding to complementary sequences in the noncoding regions of mRNA molecules transcribed from other heterochronic genes, thereby repressing their translation and promoting their degradation. Increasing levels of Lin4 miRNA govern the progression from first-stage larva cell behaviors to third-stage larva cell behaviors Increasing levels of Let7 miRNA govern the progression from late larva to adult. In fact, Lin4 and Let7 were the first miRNAs to be described in any animal: it was through developmental genetic studies in C. elegans that the importance of this whole class of molecules for gene regulation in animals was discovered. More generally, in many animals, miRNAs help regulate the transitions between different stages of development. For example, in flies, fish, and frogs, the maternal mRNAs that are loaded into the egg in the mother are removed during early development when the genome of the embryo begins to be transcribed; at this stage, the embryo begins to express specific miRNAs that target many maternal mRNAs for translational repression and degradation. Thus, miRNAs can sharpen developmental transitions by blocking and removing mRNAs that define an earlier developmental stage. But how is the timing of miRNA expression itself controlled? In the case of the miRNAs that disable maternal mRNAs in frogs and fish, expression is activated at the end of the series of rapid, synchronous divisions that cleave the fertilized egg into many smaller cells. As the division rate of these blastomeres slows, widespread transcription of the embryo’s genome begins

This event, where the embryo’s own genome largely takes over control of development from maternal macromolecules, is called the maternal-zygotic transition (MZT), and it occurs with roughly similar timing in most animal species, with the exception of mammals. One trigger for the MZT appears to be the nuclear-to-cytoplasmic ratio. During cleavage, the total amount of cytoplasm in the embryo remains constant, but the number of cell nuclei increases exponentially. As a critical threshold is reached in the ratio of cytoplasm to DNA, the cell cycles lengthen and transcription is initiated. Thus, haploid embryos undergo the MZT one cell cycle later than diploid embryos, which contain twice as much DNA per cell. According to one
Although signaling between cells plays an essential part in driving the progress of development, this does not mean that cells always need signals from other cells to prod them into changing their character as development proceeds. Some of these changes are intrinsic to the cell (like the ticking of the segmentation clock) and depend on intracellular developmental programs that can operate even when the cell is removed from its normal environment. The best-understood example is in the development of neural precursor cells, or neuroblasts, in the embryonic Drosophila central nervous system. These cells, as we saw, are initially singled out from the neurogenic ectoderm of the embryo by a typical lateral-inhibition mechanism that depends on Notch, and they then proceed through an entirely predictable series of asymmetric cell divisions to generate ganglion mother cells that divide to form neurons and glial cells.

Neuroblasts and asymmetric cell division in the central nervous system of a fly embryo. The neuroblast originates as a specialized ectodermal cell. It is singled out by lateral inhibition and emerges from the basal (internal) face of the ectoderm. It then goes through repeated division cycles, dividing asymmetrically to generate a series of ganglion mother cells. Each ganglion mother cell divides just once to give a pair of differentiated daughters (typically a neuron plus a glial cell).
The neuroblast changes its internal state as it goes through its set program of divisions, generating different cell types with a reproducible sequence and timing. These successive changes in neuroblast specification occur through the sequential expression of specific transcription regulators. For example, most embryonic neuroblasts sequentially express the transcription regulators Hunchback, Krüppel, Pdm, and Cas in a fixed order

Temporal patterning of neuroblast fate in Drosophila. Hunchback, Krüppel, Pdm, and Cas are transcription regulators that are expressed consecutively in the cell lineage of neuroblasts during development of the Drosophila nervous system. At successive time steps, correlated with cell division, the neuroblast switches its pattern of gene expression. Each neuroblast division produces one daughter that remains a neuroblast and expresses the updated set of genes, and one ganglion mother cell that maintains the expression of this gene set and differentiates into specific cell types accordingly.
When a neuroblast divides, the set of transcription regulators expressed at that time is inherited by the ganglion mother cell and its neural progeny; thus, the differentiated neural cells are endowed with different characters according to their time of birth. Remarkably, when neuroblasts are taken from an embryo and maintained in culture, isolated from their normal surroundings, they step through much the same stereotyped developmental program as if they had been left in the embryo. Moreover, many of the neuroblast transitions occur even when cell division is blocked. The neuroblasts seem to have a built-in timer how was this timer programmed ? that determines when each of the transcription regulators is expressed, and this timer can continue to run in the absence of cell division. The molecular basis of the timing is largely unknown;
in part, at least, it must depend on the time taken for gene switching, as described above; but it may well also depend on slow progressive changes in chromatin structure. These too can serve to measure the passage of time in the embryo.
Cells Rarely Count Cell Divisions to Time Their Development
Many specialized cells in animals develop from proliferating progenitor cells that stop dividing and terminally differentiate after a limited number of cell divisions. In these cases, differentiation is coordinated how was this coordination programmed ? with withdrawal from the cell cycle, but it is usually not known how the coordination is achieved. It has often been suggested that the cell-division cycle might serve as an intracellular timer to control the timing of cell differentiation. The cell cycle would be the ticking clock that sets the tempo of other developmental processes, with maturational changes in gene expression being dependent on cell-cycle progression. Most of the evidence, however, indicates that this tempting idea is wrong. Although there are examples where cells change their maturation state with each division and the change depends on cell division, this is not the general rule. As we just saw for neuroblasts in the Drosophila embryo, cells in developing animals often carry on with their normal timetable of maturation and differentiation even when cell division is artificially blocked; necessarily, some abnormalities occur, if only because a single undivided cell cannot differentiate in two ways at once. But it seems that most developing cells can change their state without a requirement for cell division. Developmental control genes can switch the cell-division-cycle machinery on or off, and it is the dynamics of these genes, rather than the cell cycle, that sets the tempo of development.
MicroRNAs Often Regulate Developmental Transitions
Genetic screens are useful for tracking down the genes involved in almost any biological process, and they have been used to search for mutations that alter developmental
timing. Such screens were performed in the nematode Caenorhabditis elegans

This worm is small, relatively simple, and precisely structured. The anatomy of its development is highly predictable and has been described in extraordinary detail, so that one can map out the exact lineage of every cell in the body and see exactly how the developmental program is altered in a mutant. Genetic screens in C. elegans revealed mutations that disrupt developmental timing in a particularly striking way: in these so-called heterochronic mutants, certain cells in a larva at one stage of development behave as though they were in a larva at a different stage of development, or cells in the adult carry on dividing as though they belonged to a larva

Genetic analyses showed that the products of the heterochronic genes act in series, forming regulatory cascades. Unexpectedly, two genes at the top of their respective cascades, called Lin4 and Let7, were found to code not for protein but instead for microRNAs (miRNAs)—short, untranslated, regulatory RNA molecules, 21 or 22 nucleotides long. These act by binding to complementary sequences in the noncoding regions of mRNA molecules transcribed from other heterochronic genes, thereby repressing their translation and promoting their degradation. Increasing levels of Lin4 miRNA govern the progression from first-stage larva cell behaviors to third-stage larva cell behaviors Increasing levels of Let7 miRNA govern the progression from late larva to adult. In fact, Lin4 and Let7 were the first miRNAs to be described in any animal: it was through developmental genetic studies in C. elegans that the importance of this whole class of molecules for gene regulation in animals was discovered. More generally, in many animals, miRNAs help regulate the transitions between different stages of development. For example, in flies, fish, and frogs, the maternal mRNAs that are loaded into the egg in the mother are removed during early development when the genome of the embryo begins to be transcribed; at this stage, the embryo begins to express specific miRNAs that target many maternal mRNAs for translational repression and degradation. Thus, miRNAs can sharpen developmental transitions by blocking and removing mRNAs that define an earlier developmental stage. But how is the timing of miRNA expression itself controlled? In the case of the miRNAs that disable maternal mRNAs in frogs and fish, expression is activated at the end of the series of rapid, synchronous divisions that cleave the fertilized egg into many smaller cells. As the division rate of these blastomeres slows, widespread transcription of the embryo’s genome begins

This event, where the embryo’s own genome largely takes over control of development from maternal macromolecules, is called the maternal-zygotic transition (MZT), and it occurs with roughly similar timing in most animal species, with the exception of mammals. One trigger for the MZT appears to be the nuclear-to-cytoplasmic ratio. During cleavage, the total amount of cytoplasm in the embryo remains constant, but the number of cell nuclei increases exponentially. As a critical threshold is reached in the ratio of cytoplasm to DNA, the cell cycles lengthen and transcription is initiated. Thus, haploid embryos undergo the MZT one cell cycle later than diploid embryos, which contain twice as much DNA per cell. According to one

