Defining the LUCA: What might be a Cell’s minimal requirement of parts? I have gone in-depth to elucidate this question in my previous book: On the Origin of Life and Virus World by means of an Intelligent Designer. I wrote: Science remains largely in the dark when it comes to pinpointing what exactly the first life form looked like. Speculation abounds. Whatever science paper about the topic one reads, confusion becomes apparent. Patrick Forterre wrote in a science paper in 2015: The universal tree of life: an update, confessed:
There is no protein or groups of proteins that can give the real species tree, i.e., allow us to recapitulate safely the exact path of life evolution.
As such, whatever architecture one comes up with, remains in the realm of speculation. Is it therefore futile, to trace a borderline, and list a number of features, that most likely were present? No. Even if we can come up only with a hypothetical organism, it will nonetheless give us insight into the complexity involved, and bring us closer to deciding, what mechanisms most likely were involved and if intelligence was required to set up the first life forms.
Andrew J. Crapitto (2022): The availability of genomic and proteomic data from across the tree of life has made it possible to infer features of the genome and proteome of the last universal common ancestor (LUCA). Several studies have done so, all using a unique set of methods and bioinformatics databases.
No individual study shares a high or even moderate degree of similarity with any other individual study. Studies of the genome or proteome of the LUCA do not uniformly agree with one another. The set of consensus LUCA protein family predictions between all of these studies portrays a LUCA genome that, at minimum, encoded functions related to protein synthesis, amino acid metabolism, nucleotide metabolism, and the use of common, nucleotide-derived organic cofactors.
The translation process is well known to be ancient and many of the proteins involved in translation machinery appear to predate the LUCA. A corollary to the influential RNA world hypothesis is that the translation system evolved within the context of an RNA-based genetic system. Most universal Clusters of Orthologous ( Orthologous are homologous genes where a gene diverges after a speciation event, but the gene and its main function are conserved) Groups of proteins (COGs) COGs encode proteins that physically associate with the ribosome and those that do not are often involved with the translation process in some other way. Similarly, nearly all universal, vertically inherited functional RNAs (save the SRP RNA) are involved in the translation system. Translation-related genes or proteins are prevalent in the predictions of seven of the eight previously published LUCA genome or proteome studies analyzed here.
We identified 366 eggNOG clusters that were predicted by four or more studies to have been present in the genome of the LUCA (Appendix S2). 7
William Martin and colleagues from the University Düsseldorf’s Institute of Molecular Evolution give us also an interesting number: The metabolism of cells contains evidence reflecting the process by which they arose. Here, we have identified the ancient core of autotrophic metabolism encompassing
404 reactions that comprise the reaction network from H2, CO2, and ammonia (NH3) to amino acids, nucleic acid monomers, and the 19 cofactors required for their synthesis. Water is the most common reactant in the autotrophic core, indicating that the core arose in an aqueous environment. Seventy-seven core reactions involve the hydrolysis of high-energy phosphate bonds, furthermore suggesting the presence of a non-enzymatic and highly exergonic chemical reaction capable of continuously synthesizing activated phosphate bonds. CO2 is the most common carbon-containing compound in the core. An abundance of NADH and NADPH-dependent redox reactions in the autotrophic core, the central role of CO2, and the circumstance that the core’s main products are far more reduced than CO2 indicate that the core arose in a highly reducing environment. The chemical reactions of the autotrophic core suggest that it arose from H2, inorganic carbon, and NH3 in an aqueous environment marked by highly reducing and continuously far from equilibrium conditions.
Supplementary Table 1. in the paper lists all 402 metabolic reactions 11,
12John I. Glass (2006):
Mycoplasma genitalium has the smallest genome of any organism that can be grown in pure culture. It has a minimal metabolism. Consequently, its genome is expected to be a close approximation to the minimal set of genes needed to sustain bacterial life. 14
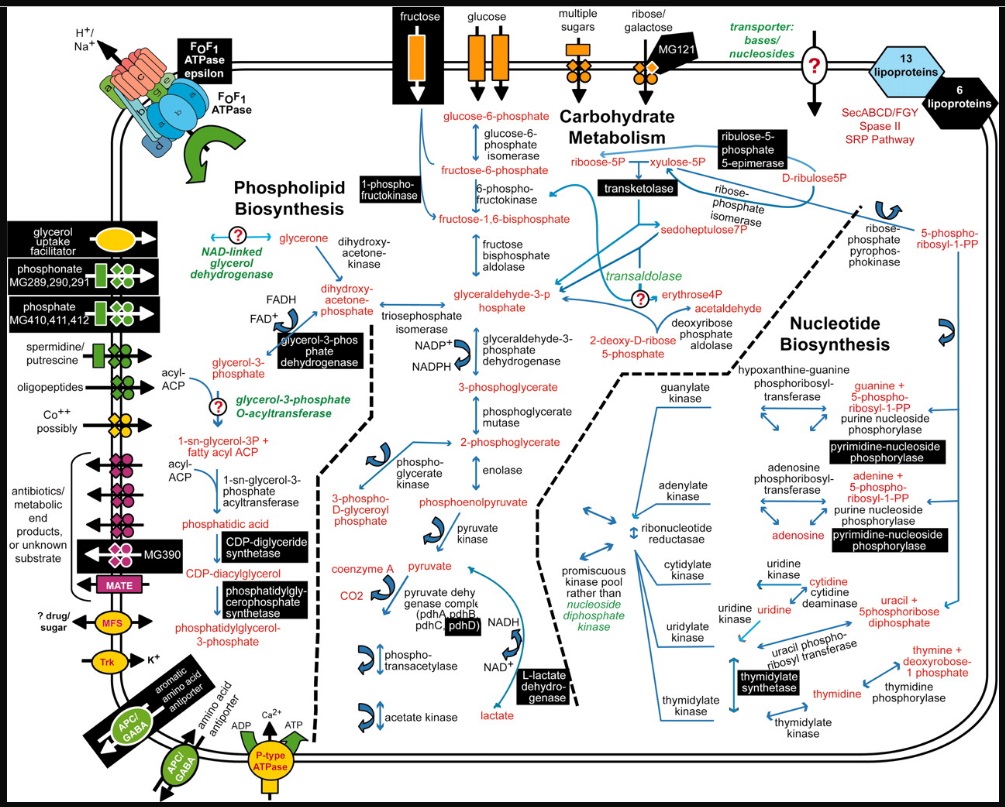 Metabolic pathways and substrate transport mechanisms encoded by M. genitalium.
Metabolic pathways and substrate transport mechanisms encoded by M. genitalium. Metabolic products are colored red, and mycoplasma proteins are black. White letters on black boxes mark nonessential functions or proteins based on our current gene disruption study. Question marks denote enzymes or transporters not identified that would be necessary to complete pathways, and those missing enzyme and transporter names are colored green. Transporters are colored according to their substrates: yellow, cations; green, anions and amino acids; orange, carbohydrates; purple, multidrug and metabolic end product efflux. The arrows indicate the predicted direction of substrate transport. The ABC type transporters are drawn as follows: rectangle, substrate-binding protein; diamonds, membrane-spanning permeases; circles, ATP-binding subunits.
J. A. G. Ranea (2006): In our view, the LUCA was faced with two important challenges associated with the source of amino acids and purine/pyrimidine bases or nucleosides. Most of these compounds need complex pathways to be synthesized and our analyses suggest that these are not present in the LUCA. Based on that, we are more in favor of amino acids and nitrogenous bases being present in a possible primitive soup rather than being synthesized by the LUCA.18
From a LUCA to the last bacterial common ancestor (LBCA)Even though the existence of LUCA is supported by the universal presence of conserved biomolecules and a vast amount of genetic data, its characteristics and identity remain unknown. LBCA, on the other hand, stands for "Last Bacterial Common Ancestor," which refers to the hypothetical ancestor of all modern bacteria. Although the characteristics of LBCA are still uncertain, recent studies suggest that it might have been a monoderm bacterium with a complete 17-gene dcw cluster, which is two genes more than in the model E. coli cluster.
The 17-gene dcw (division and cell wall) cluster is a group of bacterial genes that are involved in the regulation of cell division and the synthesis of the cell wall during the cell cycle. These genes encode proteins that are responsible for the assembly and contraction of the bacterial cell wall and septum, which eventually leads to the separation of the daughter cells. The dcw cluster includes genes that are involved in peptidoglycan synthesis, cell wall assembly, and septation, among others. These genes are found in many bacterial species and are thought to be essential for bacterial growth and survival. Understanding the composition and regulation of the dcw cluster can provide insights into bacterial cell division and the evolution of bacterial morphology.
Phylogenomic inference also reveals that the Clostridia, a class of Firmicutes, are the least diverged of the modern genomes, suggesting that the first lineage to diverge from the predicted LBCA was similar to the modern Clostridia.
In 2004, Rosario Gil proposed a minimal gene set composed of 206 genes that would sustain the main vital functions of a hypothetical simplest bacterial cell. These functions include DNA replication, transcription, translation, protein processing, folding, secretion and degradation, cell division, energetic metabolism, pentose pathway, nucleotide biosynthesis, and lipid biosynthesis. The minimal cell does not include biosynthetic pathways for amino acids or most cofactor precursors, as they can be obtained from the environment.
While some amino acids can be obtained from the environment, not all of them are readily available or in sufficient quantities to support the growth of a minimal cell. In addition, the amino acids that are available in the environment may not be in the correct proportions or forms required by the cell. Therefore, some minimal cells may require biosynthetic pathways for certain amino acids to ensure their survival and growth.
R. R. Léonard (2022): The nature of the LBCA is unknown, especially the architecture of its cell wall. The lack of reliably affiliated bacterial fossils outside Cyanobacteria makes it elusive to decide the very nature of the LBCA. Nevertheless, phylogenomic inference leads to informative results, and our analysis of the cell-wall characteristics of extant bacteria, combined with ancestral state reconstruction and distribution of key genes, opens interesting possibilities: the LBCA might have been a monoderm bacterium featuring a complete 17-gene dcw cluster, two genes more than in the model
E. coli cluster. This result was also supported by a recent study, in which the authors found 146 protein families that formed a predicted core for the metabolic network of the LBCA. From these families, phylogenetic trees were produced and the divergence of the modern genomes from the root to the tips was analysed. It appears that the
Clostridia (a class of Firmicutes) are the least diverged of the modern genomes and thus the first lineage to diverge from the predicted LBCA were similar to the modern Clostridia. Based on these results, the authors suggested that the LBCA could have been a monoderm bacteria. (Having a single membrane, especially a thick layer of peptidoglycan) 22
P. C. Morales et.al., (2019) reconstructed the phylogenetic tree of Clostridium species. They set
Clostridium difficile at the root of the tree. 23 The genome of C. difficile strain 630 consists of a circular chromosome of 4,290,252 bp 24
Taking Rosario Gil's model organism as the basis for our forthcoming investigationRosario Gil (2004): Based on the conjoint analysis of several computational and experimental strategies designed to define the minimal set of protein-coding genes that are necessary to maintain a functional bacterial cell, we propose a minimal gene set composed of 206 genes. Such a gene set will be able to sustain the main vital functions of a hypothetical simplest bacterial cell with the following features.
(i) A virtually complete DNA replication machinery, composed of one nucleoid DNA binding protein, SSB, DNA helicase, primase, gyrase, polymerase III, and ligase. No initiation and recruiting proteins seem to be essential, and the DNA gyrase is the only topoisomerase included, which should perform both replication and chromosome segregation functions.
(ii) A very rudimentary system for DNA repair, including only one endonuclease, one exonuclease, and a uracyl-DNA glycosylase.
(iii) A virtually complete transcriptional machinery, including the three subunits of the RNA polymerase, a σ factor, an RNA helicase, and four transcriptional factors (with elongation, antitermination, and transcription-translation coupling functions). Regulation of transcription does not appear to be essential in bacteria with reduced genomes, and therefore the minimal gene set does not contain any transcriptional regulators.
(iv) A nearly complete translational system. It contains the 20 aminoacyl-tRNA synthases, a methionyl-tRNA formyltransferase, five enzymes involved in tRNA maturation and modification, 50 ribosomal proteins (31 proteins for the large ribosomal subunit and 19 proteins for the small one), six proteins necessary for ribosome function and maturation (four of which are GTP binding proteins whose specific function is not well known), 12 translation factors, and 2 RNases involved in RNA degradation.
(v) Protein-processing, -folding, secretion, and degradation functions are performed by at least three proteins for posttranslational modification, two molecular chaperone systems (GroEL/S and DnaK/DnaJ/GrpE), six components of the translocase machinery (including the signal recognition particle, its receptor, the three essential components of the translocase channel, and a signal peptidase), one endopeptidase, and two proteases.
(vi) Cell division can be driven by FtsZ only, considering that, in a protected environment, the cell wall might not be necessary for cellular structure.
(vii) A basic substrate transport machinery cannot be clearly defined, based on our current knowledge. Although it appears that several cation and ABC transporters are always present in all analyzed bacteria, we have included in the minimal set only a PTS for glucose transport and a phosphate transporter. Further analysis should be performed to define a more complete set of transporters.
(viii) The energetic metabolism is based on ATP synthesis by glycolytic substrate-level phosphorylation.
(ix) The nonoxidative branch of the pentose pathway contains three enzymes (ribulose-phosphate epimerase, ribose-phosphate isomerase, and transketolase), allowing the synthesis of pentoses (PRPP) from trioses or hexoses.
(x) No biosynthetic pathways for amino acids, since we suppose that they can be provided by the environment.
(xi) Lipid biosynthesis is reduced to the biosynthesis of phosphatidylethanolamine from the glycolytic intermediate dihydroxyacetone phosphate and activated fatty acids provided by the environment.
(xii) Nucleotide biosynthesis proceeds through the salvage pathways, from PRPP and the free bases adenine, guanine, and uracil, which are obtained from the environment.
(xiii) Most cofactor precursors (i.e., vitamins) are provided by the environment. Our proposed minimal cell performs only the steps for the syntheses of the strictly necessary coenzymes tetrahydrofolate, NAD+, flavin aderine dinucleotide, thiamine diphosphate, pyridoxal phosphate, and CoA. 21
Comment: That would require LUCA to have complex import and transport mechanisms of nucleotides and amino acids, and membrane import channel proteins able to distinguish and select those building blocks for life that are used in life, from those that aren't. As I have outlined in my book, On the Origin of Life and Virus World by means of an Intelligent Designer, Origin of Life researchers have failed throughout to demonstrate the possible abiotic route to synthesize the basic building blocks of life non-enzymatically. But even IF that would be the case, that would still not explain how LUCA made the transition from external incorporation to acquire the complex metabolic and catabolic pathways to synthesize nucleotides and amino acids which constitutes a huge, often overlooked gap.
Mycoplasma genitalium is held as the smallest possible living self-replicating cell.
It is, however, a pathogen, an endosymbiont that only lives and survives within the body or cells of another organism ( humans ). As such, it IMPORTS many nutrients from the host organism. The host provides most of the nutrients such bacteria require, hence the bacteria do not need the genes for producing such compounds themselves. As such, it does not require the same complexity of biosynthesis pathways to manufacture all nutrients as a free-living bacterium. Amino Acids were no readily available on the early earth. In the Miller Urey experiment, eight of the 20 amino acids were never produced. Neither in 1953 nor in the subsequent experiments.
LUCAs information systemCurrently, there is no known form of life that exists without DNA and RNA. DNA is a fundamental component of all known life on Earth and serves as the genetic blueprint that encodes the information necessary for the development, function, and reproduction of living organisms. Some claim that it is possible that alternative forms of genetic material or information storage may exist in other environments beyond our current understanding. This is however an argument from ignorance. It is a fallacy that occurs when someone asserts a claim based on the absence of evidence to the contrary. It is important to base claims on positive evidence rather than on the absence of evidence. It is therefore warranted to start with the presumption that DNA was present when life started. And as such, as well the biosynthesis pathways necessary to synthesize deoxynucleotides, the monomer building blocks of DNA.
The Central DogmaThe term Central Dogma was coined by Francis Crick, who discovered the double-helix structure of DNA together with Rosalind Franklin, James Watson, and Maurice Wilkins.
DNA is “the Blueprint of Life.” It contains part of the data needed to make every single protein that life can't go on without. ( Epigenetic data based on epigenetic languages is also involved). No DNA, no proteins, no life. RNA has a limited coding capacity because it is unstable.
James Watson, left, with Francis Crick and their model of part of a DNA molecule
SCIENCE PHOTO LIBRARYYourGenome.org: The ‘Central Dogma’ is the process by which the instructions in DNA are converted into a functional product. It was first proposed in 1958 by Francis Crick, discoverer of the structure of DNA. The central dogma suggests that DNA contains the information needed to make all of our proteins, and that RNA is a messenger that carries this information to the ribosomes. The ribosomes serve as factories in the cell where the information is ‘translated’ from a code into a functional product. The process by which the DNA instructions are converted into the functional product is called gene expression. Gene expression has two key stages – transcription and translation. In transcription, the information in the DNA of every cell is converted into small, portable RNA messages. During translation, these messages travel from where the DNA is in the cell nucleus to the ribosomes where they are ‘read’ to make specific proteins.36
The biosynthesis of nucleotidesThe de novo biosynthesis of nucleotides is essential in cells because nucleotides serve as the building blocks of nucleic acids, which are critical for many fundamental cellular processes. Here are some key reasons why de novo nucleotide biosynthesis is essential in cells:
DNA and RNA synthesis: Nucleotides are the monomeric units that make up DNA and RNA, the two types of nucleic acids that carry genetic information in cells. De novo nucleotide biosynthesis provides the necessary raw materials for the synthesis of DNA and RNA, which are essential for cellular replication, growth, and inheritance of genetic information.
Energy storage and transfer: Nucleotides, particularly ATP (adenosine triphosphate), serve as a universal currency for energy transfer and storage in cells. ATP is used as an energy source to power numerous cellular processes, such as biosynthesis, transport of molecules across cell membranes, and cellular signaling. De novo nucleotide biosynthesis provides the precursors for the synthesis of ATP and other nucleotide-based energy molecules, which are critical for cellular energy metabolism.
Coenzymes and signaling molecules: Nucleotides also serve as important coenzymes and signaling molecules in cellular metabolism and signaling pathways. For example, NAD+ (nicotinamide adenine dinucleotide) and FAD (flavin adenine dinucleotide) are nucleotide-based coenzymes that play crucial roles in cellular redox reactions and energy metabolism. Additionally, cyclic AMP (cAMP) and GTP (guanosine triphosphate) are nucleotide-based signaling molecules that regulate various cellular processes, including cell growth, differentiation, and response to external stimuli.
Regulation of cellular processes: Nucleotides play regulatory roles in various cellular processes, such as gene expression, cell cycle progression, and immune response. For example, nucleotide-dependent enzymes, such as protein kinases and GTPases, control the activity of other proteins by phosphorylation or other post-translational modifications. Nucleotides also participate in feedback inhibition of de novo nucleotide biosynthesis, helping to regulate the cellular pool of nucleotides and maintain proper cellular nucleotide balance.
The stepwise synthesis process of nucleotides involves several key reactions and steps. Here is a general overview of the synthesis process:
1. Base synthesis: The second step is the synthesis of the nucleotide base. Bases such as adenine, guanine, cytosine, thymine, and uracil are commonly found in nucleotides. These bases can be synthesized through a variety of chemical reactions, such as the Pictet-Spengler reaction, the Fischer indole synthesis, or the Vorbrüggen glycosylation, which yield the desired base molecule.
2. Sugar moiety synthesis: The first step is the synthesis of the sugar moiety, which typically involves the formation of ribose or deoxyribose, the two common sugar molecules found in nucleotides. This can be achieved through various chemical reactions, such as the formose reaction or the Wohl degradation, which generate the desired sugar molecule.
3. Phosphate group addition: The third step is the addition of the phosphate group to the sugar molecule. This is typically achieved through phosphorylation reactions using phosphate donors, such as phosphoric acid, phosphorus oxychloride, or phosphorimidazolide. The phosphate group can be added to different positions on the sugar molecule, resulting in nucleotides with different properties and functions.
4. Nucleotide condensation: The next step is the condensation of the sugar moiety with the base and the phosphate group to form the nucleotide. This is typically achieved through chemical reactions, such as nucleophilic substitution or esterification, which result in the formation of the phosphodiester bond between the sugar and phosphate groups, with the base attached to the sugar molecule.
5. Protecting group manipulation: Throughout the synthesis process, protecting groups may be used to temporarily protect certain functional groups or prevent unwanted reactions. These protecting groups can be selectively removed or modified at specific steps using chemical reactions, allowing for the desired modifications and functionalizations of the nucleotide molecule.
6. Purification and characterization: Once the nucleotide is synthesized, it needs to be purified to remove any impurities or side products. This can be achieved through various methods, such as chromatography or crystallization. The purified nucleotide can then be characterized using techniques such as nuclear magnetic resonance (NMR) spectroscopy, mass spectrometry, or X-ray crystallography to confirm its structure and purity.
7. Further modifications: Finally, the synthesized nucleotide can be further modified or functionalized to obtain specific derivatives or analogs with desired properties or functions. This can involve additional chemical reactions, such as acylation, alkylation, or oxidation, to introduce specific functional groups or modifications to the nucleotide molecule.
We will give a closer look at what it takes to synthesize RNA and DNA. We will start with the nucleobases.
Synthesis of the RNA and DNA nucleobasesThe biosynthesis of nucleobases, which are the building blocks of nucleotides, involves complex metabolic pathways that are essential for the synthesis of RNA and DNA, the two types of nucleic acids that carry genetic information in cells.
De novo nucleobase biosynthesis: Cells can synthesize nucleobases de novo, which means starting from simple precursors and synthesizing the nucleobases from scratch. De novo nucleobase biosynthesis pathways differ for RNA and DNA, although there are some similarities. The de novo biosynthesis of nucleobases generally involves a series of enzymatic reactions that convert simple precursors into complex nucleobases through multiple intermediate steps.
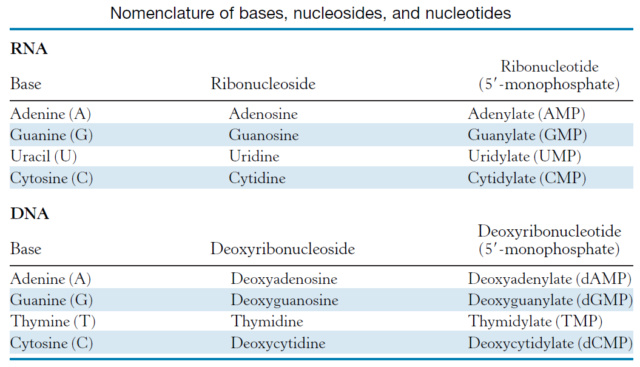
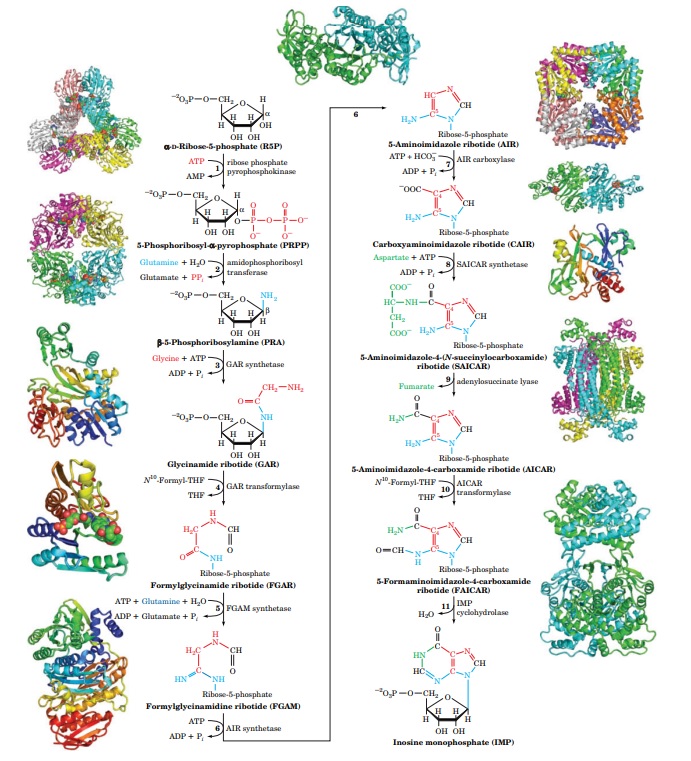
Purine nucleobase synthesis: Purine nucleobases, adenine (A) and guanine (G), are synthesized de novo from simpler precursors such as amino acids, bicarbonate, and phosphoribosyl pyrophosphate (PRPP). The biosynthesis of purine nucleobases involves several enzymatic steps, including ring construction, functional group modifications, and ring closure reactions, catalyzed by various enzymes such as amidotransferases, synthetases, and dehydrogenases.
Pyrimidine nucleobase synthesis: Pyrimidine nucleobases, cytosine (C), uracil (U), and thymine (T), are synthesized de novo from simpler precursors such as aspartate, bicarbonate, and PRPP. The biosynthesis of pyrimidine nucleobases also involves several enzymatic steps, including ring construction, functional group modifications, and ring closure reactions, catalyzed by various enzymes such as carbamoyl phosphate synthetase II (CPSII), dihydroorotase (DHOase), and orotate phosphoribosyltransferase (OPRT).
Salvage pathways: In addition to de novo biosynthesis, cells can also salvage nucleobases from the degradation of nucleic acids or from external sources, such as dietary intake. Salvage pathways involve the uptake of pre-formed nucleobases from the extracellular environment or the recycling of nucleobases from intracellular nucleotide degradation. Salvage pathways can provide an alternative source of nucleobases for nucleotide synthesis, and they are important for cellular nucleotide metabolism and conservation of resources. Salvage pathways, which involve the recycling or uptake of pre-formed nucleobases from the degradation of nucleic acids or from external sources, are not considered essential for life, as there are organisms that can survive without functional salvage pathways. However, salvage pathways play important roles in cellular nucleotide metabolism and can be advantageous for conserving resources and maintaining nucleotide pools under certain conditions.
Overall, the biosynthesis of nucleobases for RNA and DNA involves complex metabolic pathways that are essential for the synthesis of nucleotides, which are critical for the replication, transcription, and translation of genetic information in cells. De novo nucleobase biosynthesis, along with salvage pathways, ensures the availability of nucleobases for nucleotide synthesis, and proper regulation of these pathways is crucial for maintaining cellular nucleotide balance and function.
Here is a simplified overview of the minimum number of enzymes typically involved in the de novo biosynthesis of the four nucleobases used in genes (adenine, cytosine, guanine, and uracil) in most organisms:
Adenine (A) biosynthesis: The shortest pathway involves
5 enzymes: glutamine phosphoribosylpyrophosphate amidotransferase (GPAT), phosphoribosylaminoimidazole carboxamide formyltransferase (AICAR Tfase), phosphoribosylaminoimidazole succinocarboxamide synthetase (SAICAR synthetase), adenylosuccinate synthetase (ADSS), and adenylosuccinate lyase (ADSL).
Cytosine (C) biosynthesis: The shortest pathway involves
3 enzymes: carbamoyl phosphate synthetase II (CPSII), aspartate transcarbamylase (ATCase), and dihydroorotase (DHOase).
Guanine (G) biosynthesis: The shortest pathway involves
4 enzymes: inosine monophosphate (IMP) dehydrogenase (IMPDH), GMP synthase (GMPS), xanthosine monophosphate (XMP) aminase, and GMP reductase.
Uracil (U) biosynthesis: The shortest pathway involves
3 enzymes: carbamoyl phosphate synthetase II (CPSII), dihydroorotase (DHOase), and uracil phosphoribosyltransferase (UPRT).
These are simplified pathways and the actual biosynthesis of nucleobases in living organisms can be more complex, involving regulation, feedback mechanisms, and additional enzymes or intermediates. The specific enzymes and pathways for nucleobase biosynthesis can also vary depending on the organism, as different organisms may have different metabolic pathways for nucleotide biosynthesis. Regulation, feedback mechanisms, and additional enzymes or intermediates play important roles in nucleotide synthesis, as they help to maintain proper control and balance in the production of nucleotides in living organisms. While they may not be absolutely essential for nucleotide synthesis to occur, they are crucial for ensuring that nucleotide production is regulated and optimized for the needs of the cell or organism. Here's a brief overview:
Regulation: Nucleotide synthesis is typically regulated at multiple levels to maintain proper control over the production of nucleotides. Enzymes involved in nucleotide synthesis are often regulated through feedback inhibition, where the end products of nucleotide metabolism (i.e., nucleotides or their derivatives) act as feedback inhibitors, binding to specific enzymes in the synthesis pathway and inhibiting their activity. This helps to prevent overproduction of nucleotides and maintain a balanced pool of nucleotides in the cell.
Feedback mechanisms: Feedback mechanisms involve the sensing of intracellular nucleotide levels and subsequent regulation of nucleotide synthesis. For example, if the cell has sufficient nucleotide levels, feedback mechanisms may downregulate the activity of enzymes involved in nucleotide synthesis to prevent overproduction. Conversely, if nucleotide levels are low, feedback mechanisms may upregulate the activity of enzymes involved in nucleotide synthesis to meet the cellular demand.
Additional enzymes or intermediates: Nucleotide synthesis pathways often require multiple enzymes and intermediates to catalyze the various chemical reactions involved. These enzymes and intermediates may be essential for the proper progression of the synthesis pathway and the efficient production of nucleotides. For example, enzymes such as kinases, phosphatases, and ligases may be required for the addition or removal of phosphate groups during nucleotide synthesis, while intermediates such as PRPP (5-phosphoribosyl-1-pyrophosphate) may serve as critical precursors for nucleotide biosynthesis.
Here are some examples of enzymes that are involved in the regulation, feedback mechanisms, and additional intermediates of nucleotide synthesis, and are essential for the survival of the cell:
Ribonucleotide reductase: Ribonucleotide reductase is a key enzyme involved in the synthesis of deoxyribonucleotides, which are the building blocks of DNA. It catalyzes the conversion of ribonucleotides to deoxyribonucleotides, a crucial step in DNA synthesis. Ribonucleotide reductase is tightly regulated through allosteric feedback inhibition by the end products of the deoxyribonucleotide pathway, such as dATP, dGTP, dCTP, and dTTP, which bind to specific regulatory sites on the enzyme and inhibit its activity. This feedback inhibition helps to prevent overproduction of deoxyribonucleotides and maintains a balanced pool of nucleotides for DNA synthesis.
Purine and pyrimidine biosynthetic enzymes: Enzymes involved in the de novo biosynthesis of purine and pyrimidine nucleotides, such as phosphoribosyl pyrophosphate (PRPP) synthetase, adenylosuccinate synthase, and dihydroorotate dehydrogenase, are essential for nucleotide synthesis. These enzymes are regulated through feedback inhibition by the end products of the respective pathways, such as AMP, GMP, CMP, and UMP, which act as feedback inhibitors and help to maintain proper control over purine and pyrimidine nucleotide production.
Salvage pathway enzymes: Cells also have salvage pathways for recycling and salvaging nucleotides from cellular waste or exogenous sources. Enzymes involved in salvage pathways, such as hypoxanthine-guanine phosphoribosyltransferase (HGPRT) and thymidine kinase, are essential for salvaging and recycling nucleotides, as they help to replenish the cellular nucleotide pool and prevent nucleotide depletion. These salvage pathway enzymes are also regulated through feedback inhibition by the end products of nucleotide metabolism, which helps to regulate their activity and maintain nucleotide homeostasis.
Phosphatases and kinases: Enzymes such as nucleoside diphosphate kinases (NDPK), nucleotide monophosphate kinases (NMPK), and nucleotide diphosphatases (NDPases) are involved in the interconversion of nucleotide monophosphates, diphosphates, and triphosphates, and are essential for maintaining the proper balance of nucleotide pools in the cell. These enzymes are also regulated through feedback mechanisms and are important for regulating the cellular levels of nucleotide phosphates.
Enzymes involved in protecting group manipulations: Protecting groups are often used in nucleotide synthesis to temporarily protect specific functional groups or prevent unwanted reactions. Enzymes such as esterases or deprotecting enzymes are often used to selectively remove protecting groups at specific steps in the synthesis process, allowing for the desired modifications and functionalizations of the nucleotide molecule.
These are just a few examples of enzymes that are involved in the regulation, feedback mechanisms, and additional intermediates of nucleotide synthesis, and are essential for the survival of the cell. The specific enzymes and mechanisms involved may vary depending on the organism and the type of nucleotide being synthesized, but overall, these regulatory processes and enzymes are critical for maintaining proper control, balance, and efficiency in nucleotide synthesis, which is essential for cellular function and survival.
The biosynthesis of nucleobases is a complex process involving
multiple distinct biosynthetic pathways. In total, six different biosynthetic pathways are involved in the de novo synthesis of the five nucleobases that make up DNA and RNA. Adenine and guanine are derived from the purine biosynthetic pathway, which involves
10 enzymatic steps. This pathway starts with simple precursors such as glycine, glutamine, aspartate, and CO2, and involves multiple intermediate compounds such as IMP, AMP, and GMP.
Uracil, thymine, and cytosine, on the other hand, are derived from the pyrimidine biosynthetic pathway, which involves
six enzymatic steps. This pathway starts with simple precursors such as aspartate and carbamoyl phosphate, and involves intermediate compounds such as UMP, TMP, and CMP.
It's worth noting that some organisms have salvage pathways that can
recycle pre-existing nucleobases to avoid the de novo synthesis of nucleobases altogether. However, the de novo synthesis of nucleobases remains a crucial process in many organisms.
The precursors for nucleotides are largely derived from amino acids, specifically glycine and aspartate, which serve as the scaffolds for the ring systems present in nucleotides. In addition, aspartate and glutamine serve as sources of NH2 groups in nucleotide formation. In de novo pathways, pyrimidine bases are assembled first from simpler compounds and then attached to ribose.
What does de novo mean?In biochemistry, a de novo pathway is a metabolic pathway that synthesizes complex molecules from simple precursors. In other words, it is a process of creating new molecules from scratch rather than from pre-existing molecules.
De novo pathways are important for the synthesis of essential biomolecules such as nucleotides, amino acids, and fatty acids. For example, the de novo synthesis of purines and pyrimidines, the building blocks of DNA and RNA, are crucial for cell growth and replication. The term "de novo" comes from the Latin phrase "from the beginning," which reflects the fact that these pathways start with simple precursors and build up to more complex molecules through a series of biochemical reactions.
Purine bases, on the other hand, are synthesized piece by piece directly onto a ribose-based structure. These pathways consist of a small number of elementary reactions that are repeated with variations to generate different nucleotides. The simpler compounds used in the de novo pathways for nucleotide biosynthesis include carbon dioxide, amino acids (such as glycine, aspartate, and glutamine), tetrahydrofolate derivatives, ATP, and various cofactors such as NAD, NADP, and pyridoxal phosphate. The derivatives of tetrahydrofolate (THF) that are involved as cofactors in various reactions include
N10-formyl-THF,
N5,
N10-methylene-THF,N5-formimino-THF, and N5-methyl-THF. These THF derivatives
play crucial roles in providing one-carbon units for the synthesis of nucleotide bases.
One-carbon units One-carbon units are necessary for the construction of nucleotides because they are used as building blocks for the synthesis of the nitrogen-containing bases that make up the nucleotides. The nitrogen-containing bases of nucleotides, such as purines and pyrimidines, are synthesized through a
series of enzymatic reactions that involve the transfer of one-carbon units, such as formyl, methyl, methylene, and formimino groups. Formyl is a functional group consisting of a carbon atom double-bonded to an oxygen atom and single-bonded to a hydrogen atom, and its formula is -CHO. Methyl is a one-carbon unit (-CH3) used in nucleotide biosynthesis and other metabolic processes. Methylene is a functional group consisting of a carbon atom with two hydrogen atoms attached to it (-CH2-), which is present in many important compounds and is a building block in the synthesis of many organic compounds. Formimino is a functional group consisting of a nitrogen atom attached to a carbon atom double-bonded to an oxygen atom, and it is an important intermediate in various biochemical reactions, including the metabolism of amino acids and the biosynthesis of some neurotransmitters.
These one-carbon units are derived from various sources, including amino acids, carbon dioxide, and folate derivatives, and are incorporated into the nitrogen-containing rings of the nucleotide bases. For example, in the de novo synthesis of purine nucleotides, the carbon atoms for the C4, C5, and N7 atoms of the purine ring are derived from N10-formyl-THF, N5, N10-methylene-THF, and N5-formimino-THF, respectively.
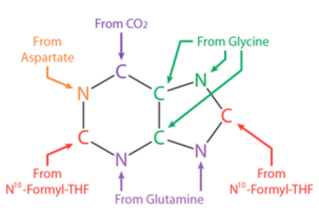
In the de novo synthesis of thymidine nucleotides, the carbon atoms for the methyl group of thymine are derived from N5, N10-methylene-THF. The biosynthesis of nucleotides is therefore closely linked to the metabolism of folate, and deficiencies in folate intake or metabolism can lead to impaired nucleotide synthesis and various pathologies.
These compounds are assembled and converted into the nucleotide bases through a series of enzymatic reactions. For example, in the de novo pathway for pyrimidine biosynthesis, carbamoyl phosphate and aspartate are condensed to form the pyrimidine ring, which is then further modified to yield uridine monophosphate (UMP). In the de novo pathway for purine biosynthesis, the purine ring is assembled stepwise onto the ribose scaffold through a series of enzyme-catalyzed reactions that utilize a variety of simpler compounds as substrates.
L. Stryer (2002): Purines and pyrimidines are derived largely from amino acids. The amino acids glycine and aspartate are the scaffolds on which the ring systems present in nucleotides are assembled. Furthermore, aspartate and the side chain of glutamine serve as sources of NH2 groups in the formation of nucleotides. In de novo (from scratch) pathways, the nucleotide bases are assembled from simpler compounds. The framework for a pyrimidine base is assembled first and then attached to ribose. In contrast, the framework for a purine base is synthesized piece by piece directly onto a ribose-based structure. These pathways each comprise a small number of elementary reactions that are repeated with variations to generate different nucleotides.53The biosynthesis of glycine, one of the two amino acids required to assemble the ring systems of nucleotides, can occur through the serine hydroxymethyltransferase (SHMT) pathway or the glycine cleavage system. The biosynthesis of aspartate, the other amino acid required to assemble the ring systems of nucleotides, can occur through the transamination of oxaloacetate.
The biosynthesis of amino acids requires a series of enzymatic reactions that convert simple molecules such as glucose or other central metabolites into the final amino acid product. These pathways are highly regulated and often require energy input from ATP or other high-energy molecules.
The serine hydroxymethyltransferase (SHMT) pathway is a biosynthetic pathway that involves the interconversion of serine and glycine, two amino acids that are important building blocks for proteins and nucleotides. In this pathway, serine is converted into glycine through the action of the enzyme serine hydroxymethyltransferase (SHMT). This enzyme transfers a methyl group from serine to tetrahydrofolate (THF), a cofactor derived from folate, and produces glycine and 5,10-methylene-THF. The SHMT pathway is important for the biosynthesis of nucleotides, which are the building blocks of DNA and RNA. In this context, the glycine produced by the SHMT pathway can be used to synthesize purines, one of the two types of nucleotide bases. Additionally, the 5,10-methylene-THF produced by the pathway can be used to produce thymidylate, a precursor for the other type of nucleotide base, pyrimidines.
The starting molecules or substrates involved in the biosynthesis pathway of the serine hydroxymethyltransferase (SHMT) pathway are serine and tetrahydrofolate (THF).
Serine is an amino acid that is used in the biosynthesis of proteins. It has a hydroxyl group (-OH) attached to its side chain and is one of the 20 common amino acids found in proteins. In addition to its role in protein synthesis, serine is also involved in the biosynthesis of other molecules such as purines, pyrimidines, and phospholipids. The biosynthesis of serine involves three enzymatic steps, which are catalyzed by
3-phosphoglycerate dehydrogenase,
phosphoserine phosphatase, and
phosphoserine aminotransferase. The biosynthesis pathways for nucleotides, including the synthesis of serine, do require enzymes. And in turn, these enzymes are encoded by genes that are themselves made of DNA. So, in a sense, DNA is required to make the enzymes that are necessary for its own biosynthesis. This is one example of how the various components of a living system are interdependent and interconnected. This interdependence between biosynthetic pathways means that the cell must maintain a delicate balance of metabolic processes to function properly. The cell achieves this balance through a complex network of biochemical reactions and regulatory mechanisms.
These reactions are finely tuned to ensure that the concentrations of various molecules are maintained within a narrow range, and that they are produced and consumed at the appropriate rates. Regulatory mechanisms, such as feedback inhibition and gene regulation, help to maintain this balance by controlling the expression and activity of enzymes involved in these pathways. Additionally, the cell has mechanisms for recycling and salvaging molecules, which helps to minimize waste and ensure that essential molecules are available for biosynthesis. Overall, the cell is able to achieve a dynamic balance through the integration of these complex biochemical and regulatory mechanisms. Maintaining the balance of biochemical reactions within the cell is essential for its survival. If the balance is disrupted or unregulated, it can lead to cell death or disease. Therefore, the cell has various mechanisms in place to regulate and control the balance of its biochemical reactions. These mechanisms can involve feedback loops, enzyme regulation, and cellular signaling pathways, among others.
Some claim that the first life forms had simpler mechanisms of regulation and that more complex regulatory systems evolved over time, but there is no concrete supportive evidence for these claims.
There are several enzymes involved in the biosynthesis of tetrahydrofolate (THF), a coenzyme that plays a critical role in nucleotide synthesis and other metabolic pathways. The pathway can vary depending on the organism, but in general, it involves at least five enzymes:
GTP cyclohydrolase I (GCH1), 6-pyruvoyltetrahydropterin synthase (PTPS), dihydropteroate synthase (DHPS), dihydrofolate reductase (DHFR), and
serine hydroxymethyltransferase (SHMT). These enzymes catalyze a series of reactions that convert GTP to THF, using various cofactors and substrates along the way. Tetrahydrofolate (THF) is an essential co-factor in many biological processes, including DNA synthesis, amino acid metabolism, and nucleotide biosynthesis. Cells cannot survive without it because THF is required for the synthesis of purines, pyrimidines, and certain amino acids that are essential for cell growth and division.
As mentioned above, aspartate depends on the transamination of oxaloacetate. Transamination is a metabolic process in which an amino group (-NH2) is transferred from an amino acid to a keto acid, resulting in the formation of a new amino acid and a new keto acid. The transfer of the amino group is catalyzed by enzymes known as
transaminases or aminotransferases.In the case of the transamination of oxaloacetate, the amino group is transferred from an amino acid (usually glutamate) to oxaloacetate, resulting in the formation of aspartate and alpha-ketoglutarate. This reaction is catalyzed by the enzyme
aspartate aminotransferase. This transamination reaction is an important step in several metabolic pathways, including the biosynthesis and degradation of amino acids. For example, aspartate is a precursor for the synthesis of several other amino acids, including methionine and threonine, and alpha-ketoglutarate can enter the citric acid cycle and be used as a source of energy for the cell.
Oxaloacetate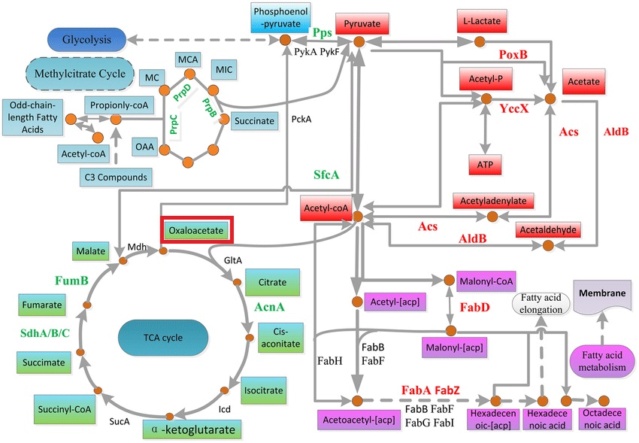
Oxaloacetate is a four-carbon dicarboxylic acid that is an important intermediate in many metabolic pathways. It is synthesized from pyruvate or other intermediates through a
series of enzymatic reactions in the mitochondrial matrix of eukaryotic cells or the cytoplasm of prokaryotic cells. One pathway for the synthesis of oxaloacetate involves the carboxylation of pyruvate, which is catalyzed by the enzyme pyruvate carboxylase. This reaction requires ATP and bicarbonate as cofactors, and results in the formation of oxaloacetate.
The complex metabolic pathways involved in the biosynthesis of the precursors to start the synthesis of nucleotides from simpler compounds demonstrate the intricate interdependence and regulation of various biochemical processes within the cell. Providing the precursors for the biosynthesis of amino acids, co-factors, and nucleotides requires a series of enzymatic reactions that are highly regulated and often require energy input from ATP or other high-energy molecules. Moreover, the biosynthesis of one molecule often depends on the availability of another molecule, resulting in a delicate balance of metabolic processes that must be maintained for the cell to function properly. This indicates that the setup is extremely unlikely to be achievable in a step-wise fashion, and an "all or nothing" approach is required, which only an intelligent designer is capable of instantiating.
The gap between the prebiotic, non-enzymatic synthesis of organic compounds and the complex metabolic pathways found in living cells is significant and multifaceted.
Prebiotic chemistry is concerned with the chemical processes that took place on Earth before the emergence of life. It is hypothesized that the basic building blocks of life, such as amino acids, nucleotides, and sugars, were formed through a series of chemical reactions that occurred spontaneously in the early Earth's environment. These reactions would have been driven by energy sources such as lightning, volcanic activity, and UV radiation.
However, the formation of these simple organic molecules would not immediately lead to the formation of complex metabolic pathways. The formation of simple organic molecules, such as amino acids and sugars, is a crucial step in the origin of life. However, the existence of these molecules alone does not lead to the formation of complex metabolic pathways. This is because the formation of metabolic pathways requires a precise interconnection of multiple enzymes, each of which performs a specific function in the pathway. Enzymes are complex protein molecules that catalyze specific chemical reactions within a cell. For a metabolic pathway to function properly, the enzymes involved in the pathway must be present in the correct sequence, with each enzyme catalyzing the correct reaction to produce the desired end product. This interconnection of enzymes is critical to the function of the pathway and requires a high degree of specificity and precision. Furthermore, the formation of enzymes is a complex process that requires a specific sequence of amino acids to fold into the correct three-dimensional structure, which is essential for its function. The probability of a random sequence of amino acids folding into a functional enzyme is extremely low, making the spontaneous formation of a functional enzyme highly unlikely. Moreover, metabolic pathways require energy to function, which must come from an external source. In modern cells, energy is provided by the breakdown of nutrients through metabolic pathways, but in the absence of such pathways, the origin of life required an external energy source. Hypothesized is the provision by geothermal energy, lightning, or radiation, among other sources. The problem here is however, these sources are very unspecific in their delivery of energy. In contrast, ATP (adenosine triphosphate) is a highly specific energy carrier that can be precisely funneled to the site of an enzyme where it is needed for a specific chemical reaction to occur.
ATP is a small molecule that is synthesized by cells through metabolic pathways, and it is used to power many cellular processes, including muscle contraction, nerve impulses, and the synthesis of molecules. ATP stores energy in its high-energy phosphate bonds, which can be released through hydrolysis to drive endergonic reactions. The specificity of ATP lies in its ability to interact with enzymes in a highly specific manner. Enzymes can bind ATP at specific sites, called active sites, which are precisely shaped to fit the ATP molecule. Once ATP is bound to an enzyme, the high-energy phosphate bond can be cleaved, releasing energy that can be used to power specific chemical reactions.
The precise delivery of ATP to the site of an enzyme is critical for its function in metabolic pathways. This is because the energy required for a specific reaction may be different from that required for another reaction in the same pathway. Therefore, the ability to funnel ATP precisely to the site where it is needed ensures that the energy is used efficiently and only where it is required.
The hypothesis of the origin of life by unguided means faces significant challenges in explaining how metabolic pathways, which rely on the highly specific energy carrier ATP, arose in the absence of modern cellular machinery. One proposed solution to this challenge is the concept of proto-metabolic pathways, which are thought to have arisen through a series of chemical reactions that were catalyzed by minerals or simple organic molecules on the early Earth. Over time, these pathways would have become more complex and interconnected, eventually leading to the emergence of metabolic pathways as we know them today.
One of the major challenges in bridging the gap between prebiotic chemistry and living organisms is the complexity of metabolic pathways found in living cells. These pathways involve a series of enzyme-catalyzed reactions that convert simple organic molecules into more complex molecules and generate the energy required for cellular functions. The origin of these pathways is claimed to have occurred over billions of years, through a process of trial and error.
This is similar to saying that: On the one side you have an intelligent agency-based system of irreducible complexity of tightly integrated, information-rich functional systems which have ready on hand energy directed for such, that routinely generate the sort of phenomenon being observed. And on the other side imagine a golfer, who has played a golf ball through a 12-hole course. Can you imagine that the ball could also play itself around the course in his absence? Of course, we could not discard, that natural forces, like wind, tornadoes, or rains or storms could produce the same result, given enough time. the chances against it, however, are so immense, that the suggestion implies that the non-living world had an innate desire to get through the 12-hole course.
The analogy of the golf ball playing itself around a course can also be applied to metabolic pathways. Metabolic pathways are complex sequences of chemical reactions that occur within cells and are responsible for the production of energy and the synthesis of various cellular components. These pathways are highly integrated, with each step depending on the previous one, and require energy to function.
Metabolic pathways require all of their parts to be present and functioning together to work. For metabolic pathways to work, all of their parts must be present and functioning together. This is because each step in the pathway is catalyzed by a specific enzyme, which is a protein that facilitates the reaction. Enzymes are highly specific in their function, meaning that each enzyme is designed to work on a specific substrate, or molecule, and produce a specific product. For example, in the process of cellular respiration, glucose is broken down into smaller molecules through a series of reactions that occur in different parts of the cell. The breakdown of glucose occurs in several stages, each catalyzed by a specific enzyme. If any one of these enzymes is missing or not functioning properly, the entire pathway is disrupted and the cell cannot produce energy efficiently. Moreover, metabolic pathways are regulated by feedback mechanisms that ensure that the rate of the pathway matches the needs of the cell. If any part of the pathway is disrupted, it can lead to a buildup of intermediate molecules that can be toxic to the cell. This highlights the importance of all the components being present and functioning together for the pathway to work correctly. Therefore, the presence and functioning of all the components of a metabolic pathway are essential for the proper functioning of the pathway. Any disruption or absence of any one of the components can lead to the breakdown of the entire pathway, emphasizing the requirement for a highly specific and integrated system to function properly.
The origin of ATP remains a significant challenge for the proto-metabolic pathway hypothesis, as the molecule is not readily available on the prebiotic Earth. One proposed solution to this challenge is that ATP would have been produced through abiotic reactions, such as the phosphorylation of ADP (adenosine diphosphate) in the presence of mineral catalysts. Other proposed mechanisms include the production of ATP through the metabolism of simpler molecules, such as acetyl-CoA. Acetyl-CoA however is not naturally found in the environment. It is synthesized within living organisms through various metabolic pathways. Another proposed solution is that ATP would have been produced through the use of alternative energy carriers, such as pyrophosphate, which is a less efficient but more readily available molecule that can be used to drive chemical reactions. While these proposed solutions are still subject to ongoing investigation and debate, it is clear that the origin of metabolic pathways and the production of highly specific energy carriers such as ATP remain significant, in my view, unsurmountable challenges for proposals of the origin of life by unguided means. Continued research in this field will probably shed even more evidence and light on the impossibility of the claim that life could have arisen on Earth by stochastic, non-designed events.
G. Zubay (2000): The most striking difference in the pathways to the purines and pyrimidines is the timing of ribose involvement. In de novo purine synthesis the purine ring is built on the ribose in a stepwise fashion. In pyrimidine synthesis, the nitrogen base is synthesized prior to the attachment of the ribose. In both instances, the ribose-5-phosphate is first activated by the addition of a pyrophosphate group to the C'-1 of the sugar to form phosphoribosyl pyrophosphate (PRPP). This activation facilitates the formation of the linkage between the C'-1 carbon of the ribose and the nitrogen of the purine and pyrimidine bases.54D. Penny (1999): An interesting picture of the LUCA is emerging. It was a fully
DNA and protein-based organism with extensive processing of RNA transcripts. 37 A. Hiyoshi (2011): All the self-reproducing cellular organisms so far examined have DNA as the genome.
E. V. Koonin (2012): All the difficulties and uncertainties of evolutionary reconstructions notwithstanding, parsimony analysis combined with less formal efforts on the reconstruction of the deep past of particular functional systems leaves no serious doubts that LUCA already possessed at least several hundred genes. In addition to the aforementioned “golden 100” genes involved in expression, this diverse gene complement consists of numerous metabolic enzymes, including pathways of the central energy metabolism and
the biosynthesis of nucleotides, amino acids, and some coenzymes, as well as some crucial membrane proteins, such as the subunits of the signal recognition particle (SRP) and the H+- ATPase. 36