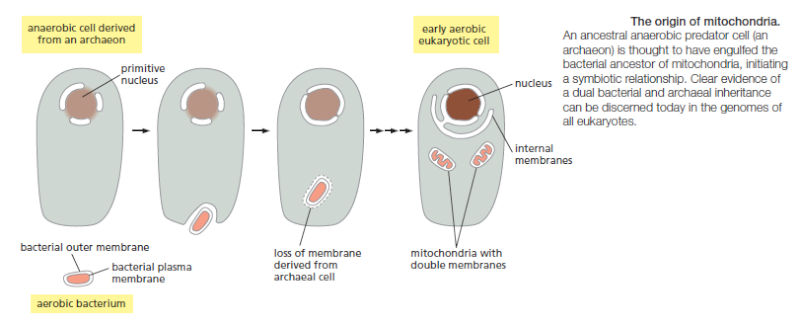
https://reasonandscience.catsboard.com/t2128-cell-membranes-origins-through-natural-mechanisms-or-design
Steven A. Benner: Paradoxes in the Origin of Life 2014
Lipids that provide tidy compartments under the close supervision of a graduate student (supporting a protocellfirst model for origins) are quite non-robust with respect to small environmental perturbations, such as a change in the salt concentration, the introduction of organic solvents, or a change in temperature.
https://www.sci-hub.ee/10.1007/s11084-014-9379-0
Commentary: In the protocell-first scenario, simple self-assembled structures like lipid bilayers are proposed as the precursors to the complex cellular membranes of living organisms. These protocells are thought to encapsulate biochemical components necessary for life, thereby creating a microenvironment conducive to biochemical reactions. However, the sensitivity of these lipid membranes to factors such as salt concentration, the presence of organic solvents, and temperature variations raises questions about their viability as stable precursors to life in the variable conditions of early Earth. For instance, a slight increase in salt concentration disrupts the integrity of the lipid bilayer, leading to the leakage of essential biomolecules. Similarly, temperature changes could affect the fluidity and permeability of the membrane, while organic solvents could dissolve the lipids, destroying the compartmentalization essential for the protocell's function. These vulnerabilities suggest that if protocells were indeed the stepping stones towards the first living cells, they would require a remarkably stable and benign environment to maintain their integrity and facilitate the transition to more complex life forms. This requirement appears to be at odds with what is known about the early Earth's environment, which was likely subject to frequent and drastic changes due to volcanic activity, meteorite impacts, and variations in temperature and chemistry. The fragility of protocells to environmental perturbations thus serves as a significant piece of evidence against the notion that such structures could spontaneously give rise to life without some form of external stabilization or guidance. This challenges the spontaneous origin of life hypothesis by underscoring the need for more robust mechanisms or conditions that could have sustained the initial steps towards life in the face of the Earth's dynamic early environment.
Some popularisers of abiogenesis like to draw diagrams showing a simple hollow sphere of lipid (a ‘vesicle’) that can form under certain conditions in a test-tube. 2 However, such a ‘membrane’ could never lead to a living cell because the cell needs to get things through the cell membrane, in both directions. Such transport into and out of the cell entails very complex protein-lipid complexes known as transport channels, which operate like electro-mechanical pumps. They are specific to the various chemicals that must pass into and out of the cell (a pump that is designed to move water will not necessarily be suitable for pumping oil). Many of these pumps use energy compounds such as ATP to actively drive the movement against the natural gradient. Even when movement is with the gradient, from high to low concentration, it is still facilitated by carrier proteins.
The cell membrane also enables a cell to maintain a stable pH, necessary for enzyme activity, and favourable concentrations of various minerals (such as not too much sodium). This requires transport channels (‘pumps’) that specifically move hydrogen ions (protons) under the control of the cell. These pumps are highly selective.10
Transport across membranes is so important that “20–30% of all genes in most genomes encode membrane proteins”.11 The smallest known genome of a free-living organism, that of the parasite Mycoplasma genitalium, codes for 26 transporters12 amongst its 482 protein-coding genes.
A pure lipid membrane would not allow even the passive movement of the positively-charged ions of mineral nutrients such as calcium, potassium, magnesium, iron, manganese, etc., or the negatively-charged ions such as phosphate, sulfate, etc., into the cell, and they are all essential for life. A pure-lipid membrane would repel such charged ions, which dissolve in water, not lipid. Indeed, a simple fat membrane would prevent the movement of water itself (try mixing a lipid like olive oil with water)!
Membrane transporters would appear to be essential for a viable living cell.
The complexity of the lipidome is often not fully appreciated. Eukaryotic cells contain phospholipids, sphingolipids, and glycolipids with a wide variation in the hydrophilic headgroups as well as diverse fatty acid compositions. Add to this complexity other lipids such as steroids and fatty acid derived cellular products, and the complexity of the lipidome equals or exceeds that of the proteome. 1
According to this website : The Interdependency of Lipid Membranes and Membrane Proteins
The cell membrane contains various types of proteins, including ion channel proteins, proton pumps, G proteins, and enzymes. These membrane proteins function cooperatively to allow ions to penetrate the lipid bilayer. The interdependency of lipid membranes and membrane proteins suggests that lipid bilayers and membrane proteins co-evolved together with membrane bioenergetics.
The nonsense of this assertion is evident. How could the membrane proteins co-evolve, if they had to be manufactured by the machinery , protected by the cell membrane ?
The cell membrane contains various types of proteins, including ion channel proteins, proton pumps, G proteins, and enzymes. These membrane proteins function cooperatively to allow ions to penetrate the lipid bilayer.
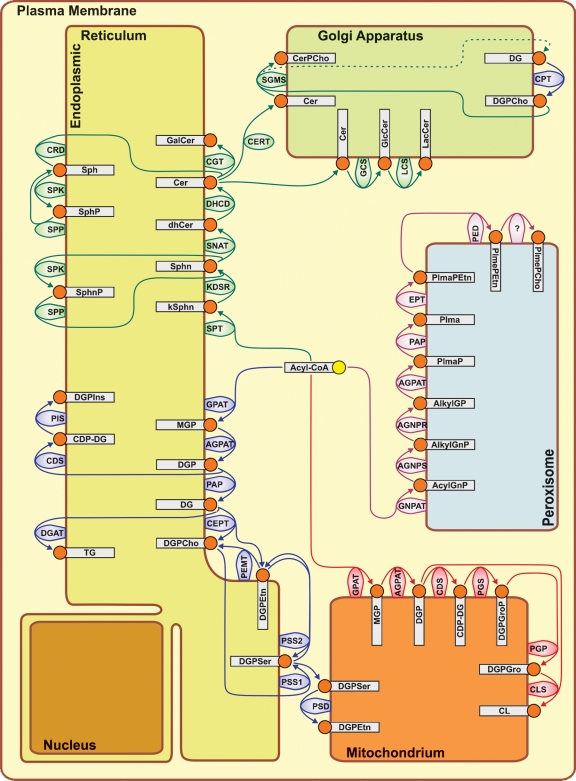
The ER and Golgi apparatus together constitute the endomembrane compartment in the cytoplasm of eukaryotic cells. The endomembrane compartment is a major site of lipid synthesis, and the ER is where not only lipids are synthesized, but membrane-bound proteins and secretory proteins are also made.
So in order to make cell membranes, the Endoplasmic Recticulum is required. But also the Golgi Apparatus, the peroxysome, and the mitochondria. But these only function, if protected and encapsulated in the cell membrane. What came first, the cell membrane, or the endoplasmic recticulum ? This is one of many other catch22 situations in the cell, which indicate that the cell could not emerge in a stepwise gradual manner, as proponents of natural mechanisms want to make us believe.
Not only is the cell membrane intricate and complex (and certainly not random), but it has tuning parameters such as the degree to which the phospholipid tails are saturated. It is another example of a sophisticated biological design about which evolutionists can only speculate. Random mutations must have luckily assembled molecular mechanisms which sense environmental challenges and respond to them by altering the phospholipid population in the membrane in just the right way. Such designs are tremendously helpful so of course they would have been preserved by natural selection. It is yet another example of how silly evolutionary theory is in light of scientific facts.
Membrane Structure
Lipids
In most cell membranes, lipids are the most abundant type of macromolecule present. Plasma and organelle membranes contain between 40% and 80% lipid. These lipids provide both the basic structure and the framework of the membrane and also regulate its function. Three types of lipids are found in cell membranes: phospholipids, cholesterol, and glycolipids.
1. Phospholipids: The most abundant of the membrane lipids are the phospholipids. They are polar, ionic compounds that are amphipathic in nature. That is, they have both hydrophilic and hydrophobic components. The hydrophilic or polar portion is in the “head group”. Within the head group is the phosphate and an alcohol that is attached to it. The alcohol can be serine, ethanolamine, inositol, or choline. Names of phospholipids then include phosphatidylserine, phosphatidylethanolamine, phosphatidylinositol, and phosphatidylcholine. While all these phospholipids contain a molecule called glycerol, the membrane phospholipid sphingomyelin has the alcohol choline in its head group and contains sphingosine instead of glycerol.
The hydrophobic portion of the phospholipid is a long, hydrocarbon (structure of carbons and hydrogens) fatty acid tail. While the polar head groups of the outer leafl et extend outward toward the environment, the fatty acid tails extend inward. Fatty acids may be saturated, containing the maximum number of hydrogen atoms bound to carbon atoms, or unsaturated with one or more carbonto- carbon double bonds. The length of the fatty acid chains and their degree of saturation impact the membrane structure. The fatty acid chains normally undergo motions such as fl exion (bending or fl exing), rotation, and lateral movement . Whenever a carbon-to-carbon double bond exists, there is a kink in the chain, reducing some types of motions and preventing the fatty acids from packing tightly together. Phospholipids in plasma membranes of healthy cells do not migrate or fl ip-fl op from one leafl et to the other. (However, during the process of programmed cell death, enzymes catalyze the movement of phosphatidylserine from the inner leaflet to the outer leaflet
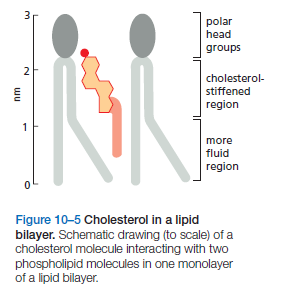
Cholesterol: Another major component of cell membranes is cholesterol. An amphipathic molecule, cholesterol contains a polar hydroxyl group as well as a hydrophobic steroid ring and attached hydrocarbon . Cholesterol is dispersed throughout cell membranes, intercalating between phospholipids. Its polar hydroxyl group is near the polar head groups of the phospholipids while the steroid ring and hydrocarbon tails of cholesterol are oriented parallel to those of the phospholipids. Cholesterol fits into the spaces created by the kinks of the unsaturated fatty acid tails, decreasing the ability of the fatty acids to undergo motion and therefore causing stiffening and strengthening of the membrane.
Glycolipids: Lipids with attached carbohydrate (sugars), glycolipids are found in cell membranes in lower concentration than phospholipids and cholesterol. The carbohydrate portion is always oriented toward the outside of the cell, projecting into the environment. Glycolipids help to form the carbohydrate coat observed on cells and are involved in cell-to-cell interactions. They are a source of blood group antigens and also can act as receptors for toxins including those from cholera and tetanus.
Proteins
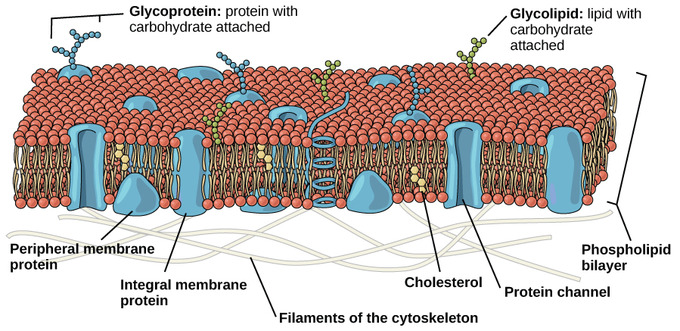
While lipids form the main structure of the membrane, proteins are largely responsible for many biological functions of the membrane. For example, some membrane proteins function in transport of materials into and out of cells. Others serve as receptors for hormones or growth factors. The types of proteins within a plasma membrane vary depending on the cell type. However, all membrane proteins are associated with membrane in one of three main ways.
Membrane associations of proteins: While some proteins span the membrane with structures that cross from one side to the other, others are anchored to membrane lipids and still others are only peripherally associated with the cytosolic side of a plasma membrane
Lipid-anchored proteins: Members of the second category of membrane proteins are lipid-anchored proteins that are attached covalently to a portion of a lipid without entering the core portion of the bilayer of the membrane. Both transmembrane and lipid-anchored proteins are integral membrane proteins since they can only be removed from a membrane by disrupting the entire membrane structure.
Peripheral membrane proteins: Proteins in the third category are peripheral membrane proteins. These proteins are located on the cytosolic side of the membrane and are only indirectly attached to the lipid of the membrane; they bind to other proteins that are attached to the lipids. Cytoskeletal proteins, such as those involved in the spectrin membrane skeleton of erythrocytes, are examples of peripheral membrane proteins
Membrane protein functions: Membrane proteins enable cells to function as members of a tissue . For example, cell adhesion molecules are proteins that extend to the surface of cells and enable cell-to-cell contact . Other membrane proteins function as ion channels and transport proteins to enable molecules to enter and exit a cell. Membrane proteins that are ligand receptors enable cells to respond to hormones and other signaling molecules. The preceding examples of membrane proteins are of integral, transmembrane proteins whose structures span the bilayer. Lipid- anchored membrane proteins include the G proteins, which are named for their ability to bind to guanosine triphosphate (GTP) and participate in cell signaling in response to certain hormones .Peripheral membrane proteins include cytoskeletal proteins that attach to the membrane and regulate its shape and stabilize its structure. Some other peripheral membrane proteins are also involved in cell signaling and include enzymes attached to the inner membrane leafl et that are activated after a hormone binds to a protein receptor
Cell membranes are crucial to the life of the cell. The plasma membrane encloses the cell, defines its boundaries, and maintains the essential differences between the cytosol and the extracellular environment. Inside eukaryotic cells, the membranes of the nucleus, endoplasmic reticulum, Golgi apparatus, mitochondria, and other membrane-enclosed organelles maintain the characteristic differences between the contents of each organelle and the cytosol. Ion gradients across membranes, established by the activities of specialized membrane proteins, can be used to synthesize ATP, to drive the transport of selected solutes across the membrane, or, as in nerve and muscle cells, to produce and transmit electrical signals. In all cells, the plasma membrane also contains proteins that act as sensors of external signals, allowing the cell to change its behavior in response to environmental cues, including signals from other cells; these protein sensors, or receptors, transfer information—rather than molecules—across the membrane. Despite their differing functions, all biological membranes have a common general structure: each is a very thin film of lipid and protein molecules, held together mainly by noncovalent interactions. Cell membranes are dynamic, fluid structures, and most of their molecules move about in the plane of the membrane. The lipid molecules are arranged as a continuous double layer about 5 nm thick.
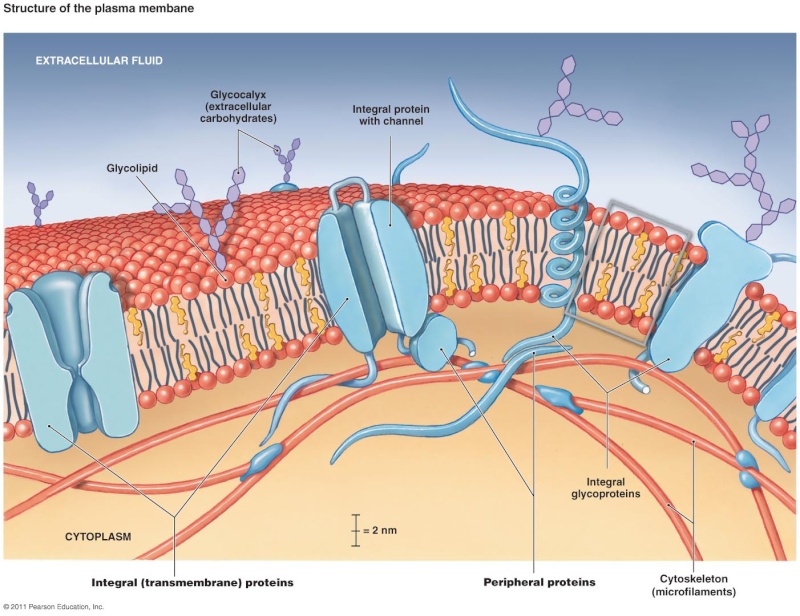
This Lipid bilayer provides the basic fluid structure of the membrane and serves as a relatively impermeable barrier to the passage of most water-soluble molecules. Most membrane proteins span the lipid bilayer and mediate nearly all of the other functions of the membrane, including the transport of specific molecules across it, and the catalysis of membrane-associated reactions such as ATP synthesis. In the plasma membrane, some transmembrane proteins serve as structural links that connect the cytoskeleton through the lipid bilayer to either the extracellular matrix or an adjacent cell, while others serve as receptors to detect and transduce chemical signals in the cell’s environment. It takes many kinds of membrane proteins to enable a cell to function and interact with its environment, and it is estimated that about 30% of the proteins encoded in an animal’s genome are membrane proteins. In this chapter, we consider the structure and organization of the two main constituents of biological membranes—the lipids and the proteins. Although we focus mainly on the plasma membrane, most concepts discussed apply to the various internal membranes of eukaryotic cells as well.
The Lipid Bilayer
The lipid bilayer provides the basic structure for all cell membranes. It is easily seen by electron microscopy, and its bilayer structure is attributable exclusively to the special properties of the lipid molecules, which assemble spontaneously into bilayers even under simple artificial conditions. In this section, we discuss the different types of lipid molecules found in cell membranes and the general properties of lipid bilayers.
Phosphoglycerides, Sphingolipids, and Sterols Are the Major Lipids in Cell Membranes
Lipid molecules constitute about 50% of the mass of most animal cell membranes, nearly all of the remainder being protein. There are approximately 5 × 106 lipid molecules in a 1 μm × 1 μm area of lipid bilayer, or about 109 lipid molecules in the plasma membrane of a small animal cell. All of the lipid molecules in cell membranes are amphiphilic—that is, they have a hydrophilic (“water-loving”) or polar end and a hydrophobic (“water-fearing”) or nonpolar end. The most abundant membrane lipids are the phospholipids. These have a polar head group containing a phosphate group and two hydrophobic hydrocarbon tails. In animal, plant, and bacterial cells, the tails are usually fatty acids, and they can differ in length (they normally contain between 14 and 24 carbon atoms). One tail typically has one or more cis-double bonds (that is, it is unsaturated), while the other tail does not (that is, it is saturated).
As shown in Figure the figure below:
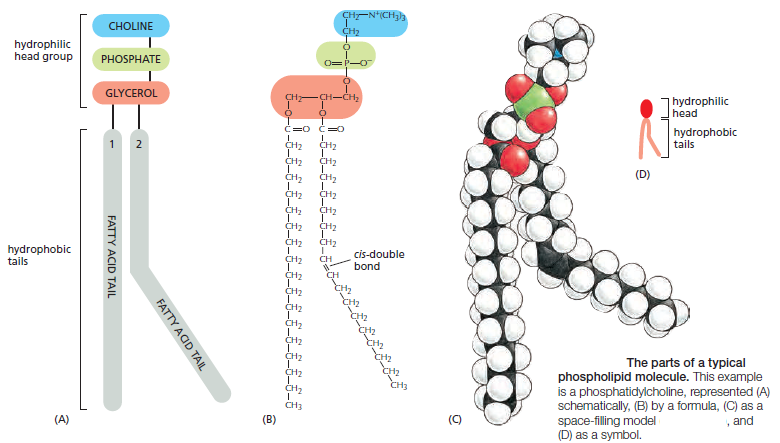
each cis-double bond creates a kink in the tail. Differences in the length and saturation of the fatty acid tails influence how phospholipid molecules pack against one another, thereby affecting the fluidity of the membrane, as we discuss later. The main phospholipids in most animal cell membranes are the phosphoglycerides, which have a three-carbon glycerol backbone Two long-chain fatty acids are linked through ester bonds to adjacent carbon atoms of the glycerol, and the third carbon atom of the glycerol is attached to a phosphate group, which in turn is linked to one of several types of head group. By combining several different fatty acids and head groups, cells make many different phosphoglycerides. Phosphatidylethanolamine, phosphatidylserine, and phosphatidylcholine are the most abundant ones in mammalian cell membranes. In addition to phospholipids, the lipid bilayers in many cell membranes contain glycolipids and cholesterol. Glycolipids resemble sphingolipids, but, insteadof a phosphate-linked head group, they have sugars attached. We discuss glycolipids later. Eukaryotic plasma membranes contain especially large amounts of cholesterol—up to one molecule for every phospholipid molecule. Cholesterol is a sterol. It contains a rigid ring structure, to which is attached a single polar hydroxyl group and a short nonpolar hydrocarbon chain
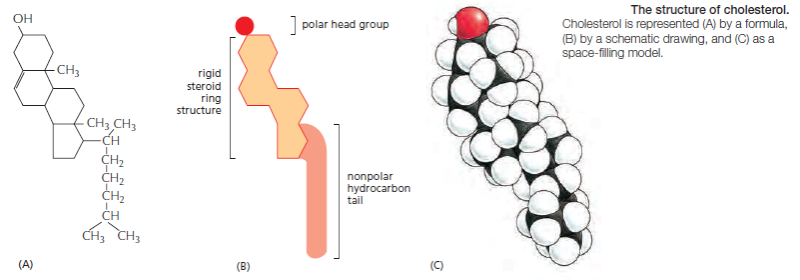
The cholesterol molecules orient themselves in the bilayer with their hydroxyl group close to the polar head groups of adjacent phospholipid molecules
Phospholipids Spontaneously Form Bilayers
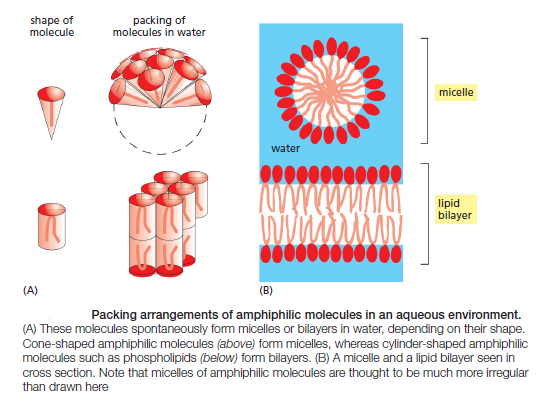
The shape and amphiphilic nature of the phospholipid molecules cause them to form bilayers spontaneously in aqueous environments. hydrophilic molecules dissolve readily in water because they contain charged groups or uncharged polar groups that can form either favorable electrostatic interactions or hydrogen bonds with water molecules
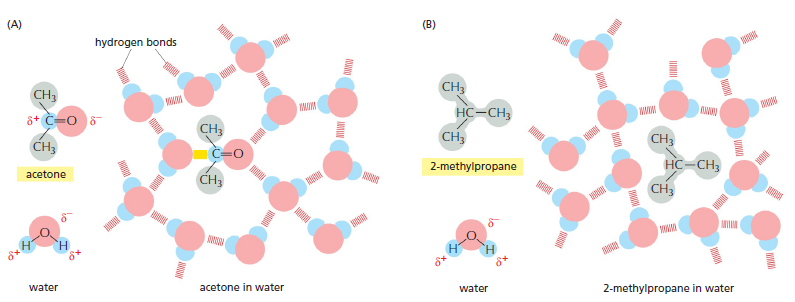
Figure above How hydrophilic and hydrophobic molecules interact differently with water. (A) Because acetone is polar, it can form hydrogen bonds (red) and favorable electrostatic interactions (yellow) with water molecules, which are also polar. Thus, acetone readily dissolves in water. (B) By contrast, 2-methyl propane is entirely hydrophobic. Because it cannot form favorable interactions with water, it forces adjacent water molecules to reorganize into icelike cage structures, which increases the free energy. This compound is therefore virtually insoluble in water. The symbol δ– indicates a partial negative charge, and δ+ indicates a partial positive charge. Polar atoms are shown in color and nonpolar groups are shown in gray.
Hydrophobic molecules, by contrast, are insoluble in water because all, or almost all, of their atoms are uncharged and nonpolar and therefore cannot form energetically favorable interactions with water molecules. If dispersed in water, they force the adjacent water molecules to reorganize into icelike cages that surround the hydrophobic molecule . Because these cage structures are more ordered than the surrounding water, their formation increases the free energy. This free-energy cost is minimized, however, if the hydrophobic molecules (or the hydrophobic portions of amphiphilic molecules) cluster together so that the smallest number of water molecules is affected. When amphiphilic molecules are exposed to an aqueous environment, they behave as you would expect from the above discussion. They spontaneously aggregate to bury their hydrophobic tails in the interior, where they are shielded from the water, and they expose their hydrophilic heads to water. Depending on their shape, they can do this in either of two ways: they can form spherical micelles, with the tails inward, or they can form double-layered sheets, or bilayers, with the hydrophobic tails sandwiched between the hydrophilic head groups.
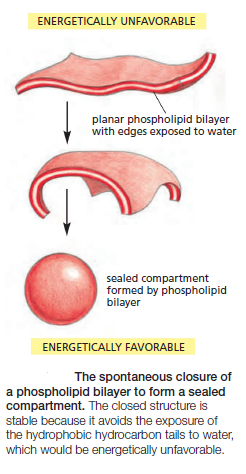
The same forces that drive phospholipids to form bilayers also provide a self-sealing property. A small tear in the bilayer creates a free edge with water; because this is energetically unfavorable, the lipids tend to rearrange spontaneously to eliminate the free edge. (In eukaryotic plasma membranes, the fusion of intracellular vesicles repairs larger tears.) The prohibition of free edges has a profound consequence: the only way for a bilayer to avoid having edges is by closing in on itself and forming a sealed compartment
This remarkable behavior, fundamental to the creation of a living cell, follows directly from the shape and amphiphilic nature of the phospholipid molecule. A lipid bilayer also has other characteristics that make it an ideal structure for cell membranes. One of the most important of these is its fluidity, which is crucial to many membrane functions
The Lipid Bilayer Is a Two-dimensional Fluid
The Fluidity of a Lipid Bilayer Depends on Its Composition
The fluidity of cell membranes has to be precisely regulated. Certain membrane transport processes and enzyme activities, for example, cease when the bilayer viscosity is experimentally increased beyond a threshold level. The fluidity of a lipid bilayer depends on both its composition and its temperature, as is readily demonstrated in studies of synthetic lipid bilayers. A synthetic bilayer made from a single type of phospholipid changes from a liquid state to a two-dimensional rigid crystalline (or gel) state at a characteristic temperature. This change of state is called a phase transition, and the temperature at which it occurs is lower (that is, the membrane becomes more difficult to freeze) if the hydrocarbon chains are short or have double bonds. A shorter chain length reduces the tendency of the hydrocarbon tails to interact with one another, in both the same and opposite monolayer, and cis-double bonds produce kinks in the chains that make them more difficult to pack together, so that the membrane remains fluid at lower temperatures
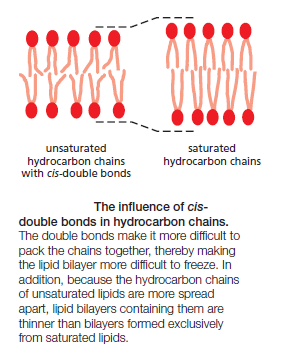
Bacteria, yeasts, and other organisms whose temperature fluctuates with that of their environment adjust the fatty acid composition of their membrane lipids to maintain a relatively constant fluidity. As the temperature falls, for instance, the cells of those organisms synthesize fatty acids with more cis-double bonds, thereby avoiding the decrease in bilayer fluidity that would otherwise result from the temperature drop. Cholesterol modulates the properties of lipid bilayers. When mixed with phospholipids, it enhances the permeability-barrier properties of the lipid bilayer. Cholesterol inserts into the bilayer with its hydroxyl group close to the polar head groups of the phospholipids, so that its rigid, platelike steroid rings interact with—and partly immobilize—those regions of the hydrocarbon chains closest to the polar head groups
By decreasing the mobility of the first few CH2 groups of the chains of the phospholipid molecules, cholesterol makes the lipid bilayer less deformable in this region and thereby decreases the permeability of the bilayer to small water-soluble molecules. Although cholesterol tightens the packing of the lipids in a bilayer, it does not make membranes any less fluid. At the high concentrations found in most eukaryotic plasma membranes, cholesterol also prevents the hydrocarbon chains from coming together and crystallizing.
The lipid composition of a typical eukaryotic cell membrane is much more complex than originally thought. These membranes contain a bewildering variety of perhaps 500–2000 different lipid species with even the simple plasma membrane of a red blood cell containing well over 150. While some of this complexity reflects the combinatorial variation in head groups, hydrocarbon chain lengths, and desaturation of the major phospholipid classes, some membranes also contain many structurally distinct minor lipids, at least some of which have important functions. The inositol phospholipids, for example, are present in small quantities in animal cell membranes and have crucial functions in guiding membrane traffic and in cell signaling
Their local synthesis and destruction are regulated by a large number of enzymes, which create both small intracellular signaling molecules and lipid docking sites on membranes that recruit specific proteins from the cytosol
Despite Their Fluidity, Lipid Bilayers Can Form Domains of Different Compositions
Because a lipid bilayer is a two-dimensional fluid, we might expect most types of lipid molecules in it to be well mixed and randomly distributed in their own monolayer. The van der Waals attractive forces between neighboring hydrocarbon tails are not selective enough to hold groups of phospholipid molecules together. With certain lipid mixtures in artificial bilayers, however, one can observe phase segregations in which specific lipids come together in separate domains
A model of a raft domain. Weak protein–protein, protein–lipid, and lipid–lipid interactions reinforce one another to partition the interacting components into raft domains. Cholesterol, sphingolipids, glycolipids, glycosylphosphatidylinositol (GPI)-anchored proteins, and some transmembrane proteins are enriched in these domains. Note that because of
their composition, raft domains have an increased membrane thickness.We discuss glycolipids, GPI-anchored proteins, and oligosaccharide linkers later.
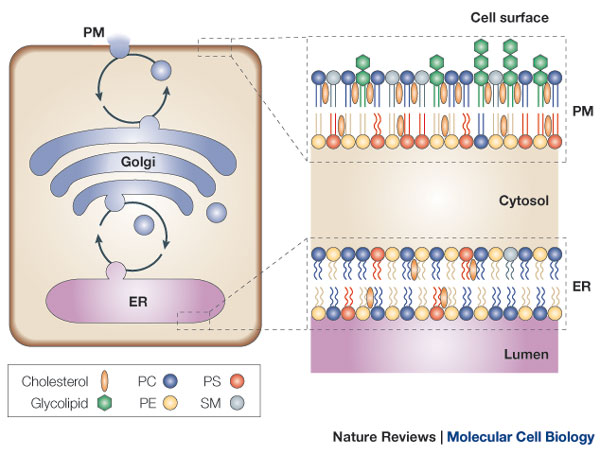
The Asymmetry of the Lipid Bilayer Is Functionally Important
The lipid compositions of the two monolayers of the lipid bilayer in many membranes are strikingly different. In the human red blood cell (erythrocyte) membrane, for example, almost all of the phospholipid molecules that have choline—( CH3)3N+CH2CH2OH—in their head group (phosphatidylcholine and sphingomyelin) are in the outer monolayer, whereas almost all that contain a terminal primary amino group (phosphatidylethanolamine and phosphatidylserine) are in the inner monolayer
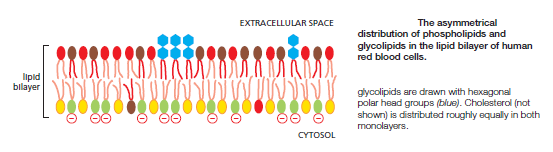
Because the negatively charged phosphatidylserine is located in the inner monolayer, there is a significant difference in charge between the two halves of the bilayer. Lipid asymmetry is functionally important, especially in converting extracellular signals into intracellular ones . Many cytosolic proteins bind to specific lipid head groups found in the cytosolic monolayer of the lipid bilayer. The enzyme protein kinase C (PKC), for example, which is activated in response to various extracellular signals, binds to the cytosolic face of the plasma membrane, where phosphatidylserine is concentrated, and requires this negatively charged phospholipid for its activity. In other cases, specific lipid head groups must first be modified to create protein-binding sites at a particular time and place. One example is phosphatidylinositol (PI), one of the minor phospholipids that are concentrated in the cytosolic monolayer of cell membranes
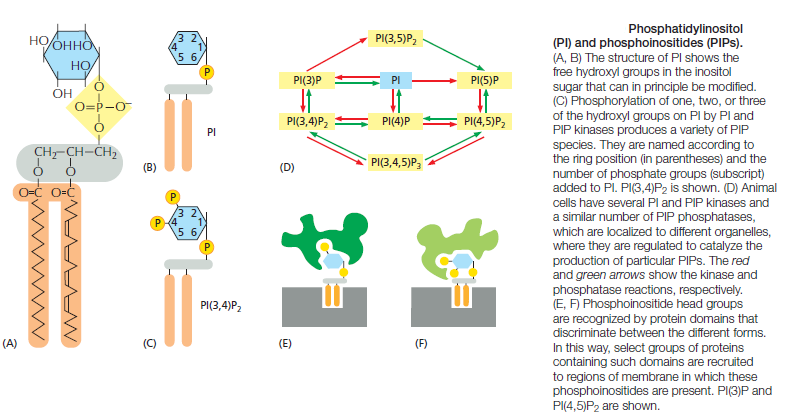
Various lipid kinases can add phosphate groups at distinct positions on the inositol ring, creating binding sites that recruit specific proteins from the cytosol to the membrane. An important example of such a lipid kinase is phosphoinositide 3-kinase (PI 3-kinase), which is activated in response to extracellular signals and helps to recruit specific intracellular signaling proteins to the cytosolic face of the plasma membrane
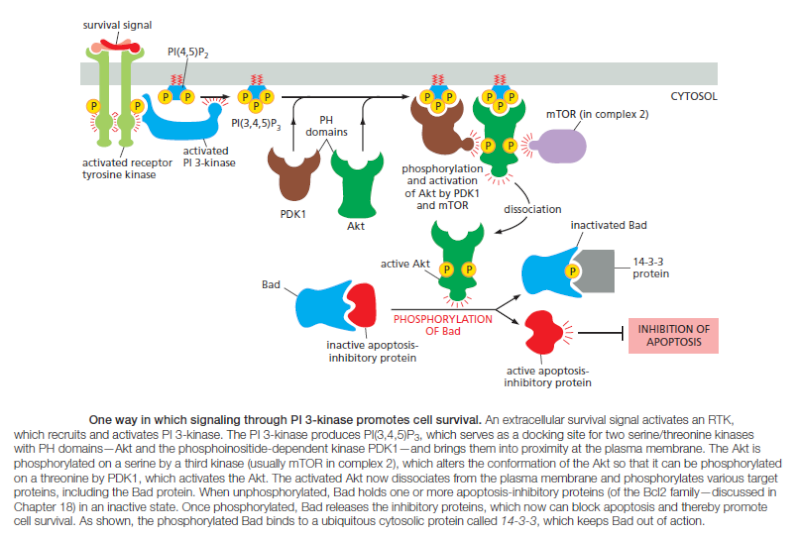
Similar lipid kinases phosphorylate inositol phospholipids in intracellular membranes and thereby help to recruit proteins that guide membrane transport. Phospholipids in the plasma membrane are used in yet another way to convert extracellular signals into intracellular ones. The plasma membrane contains various phospholipases that are activated by extracellular signals to cleave specific phospholipid molecules, generating fragments of these molecules that act as short-lived intracellular mediators. Phospholipase C, for example, cleaves an inositol phospholipid in the cytosolic monolayer of the plasma membrane to generate two fragments, one of which remains in the membrane and helps activate protein kinase C, while the other is released into the cytosol and stimulates the release of Ca2+ from the endoplasmic reticulum
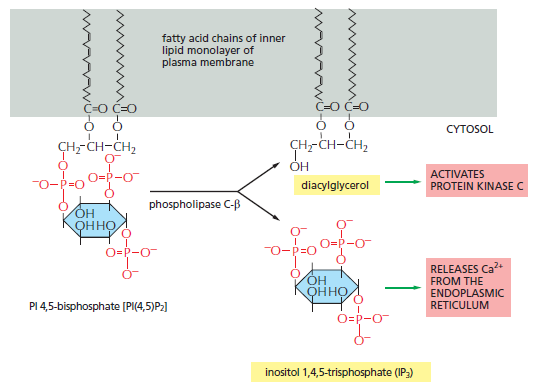
The hydrolysis of PI(4,5) P2 by phospholipase C-β. Two second messengers are produced directly from the hydrolysis of PI(4,5)P2: inositol 1,4,5-trisphosphate (IP3), which diffuses
through the cytosol and releases Ca2+ from the endoplasmic reticulum, and diacylglycerol, which remains in the membrane and helps to activate protein kinase C (PKC see below).
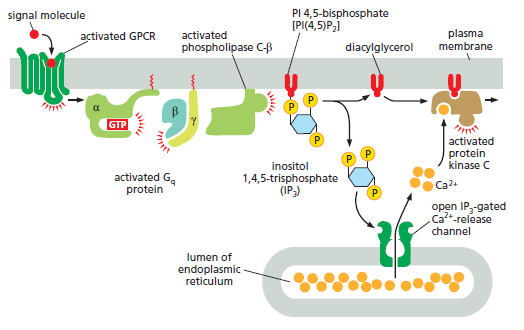
How GPCRs increase cytosolic Ca2+ and activate protein kinase C. The activated GPCR stimulates the plasma-membrane-bound phospholipase C-β (PLCβ) via a G protein called Gq. The α subunit and βγ complex of Gq are both involved in this activation. Two second messengers are produced when PI(4,5)P2 is hydrolyzed by activated PLCβ. Inositol 1,4,5-trisphosphate (IP3) diffuses through the cytosol and releases Ca2+ from the ER by binding to and opening IP3-gated Ca2+-release channels (IP3 receptors) in the ER membrane. The large electrochemical gradient for Ca2+ across this membrane causes Ca2+ to escape into the cytosol when the release channels are opened. Diacylglycerol remains in the plasma membrane and, together with phosphatidylserine (not shown) and Ca2+, helps to activate protein kinase C (PKC), which is recruited from the cytosol to the cytosolic face of the plasma membrane. Of the 10 or more distinct isoforms of PKC in humans, at least 4 are activated by diacylglycerol. There are several classes of phospholipase C: these include the β class, which is activated by GPCRs; as we see later, the γ class is activated by a class of enzymecoupled receptors called receptor tyrosine
kinases (RTKs).
Once activated, PKC phosphorylates target proteins that vary depending on the cell type. The principles are the same as discussed earlier for PKA, although most of the target proteins are different. Diacylglycerol can be further cleaved to release arachidonic acid, which can either act as a signal in its own right or be used in the synthesis of other small lipid signal molecules called eicosanoids. Most vertebrate cell types make eicosanoids, including prostaglandins, which have many biological activities. They participate in pain and inflammatory responses, for example, and many anti-inflammatory drugs (such as aspirin, ibuprofen, and cortisone) act in part by inhibiting their synthesis.
Ca2+ Functions as a Ubiquitous Intracellular Mediator
Many extracellular signals, and not just those that work via G proteins, trigger an increase in cytosolic Ca2+ concentration. In muscle cells, Ca2+ triggers contraction, and in many secretory cells, including nerve cells, it triggers secretion. Ca2+ has numerous other functions in a variety of cell types. Ca2+ is such an effective signaling mediator because its concentration in the cytosol is normally very low (~10–7 M), whereas its concentration in the extracellular fluid (~10–3 M) and in the lumen of the ER [and sarcoplasmic reticulum (SR) in muscle] is high. Thus, there is a large gradient tending to drive Ca2+ into the cytosol across both the plasma membrane and the ER or SR membrane. When a signal transiently opens Ca2+ channels in these membranes, Ca2+ rushes into the cytosol, and the resulting 10–20-fold increase in the local Ca2+ concentration activates Ca2+ responsive proteins in the cell. Some stimuli, including membrane depolarization, membrane stretch, and certain extracellular signals, activate Ca2+ channels in the plasma membrane, resulting in Ca2+ influx from outside the cell. Other signals, including the GPCR-mediated signals described earlier, act primarily through IP3 receptors to stimulate Ca2+ release from intracellular stores in the ER (see Figure 15–29). The ER membrane also contains a second type of regulated Ca2+ channel called the ryanodine receptor (so called because it is sensitive to the plant alkaloid ryanodine), which opens in response to rising Ca2+ levels and thereby amplifies the Ca2+ signal, as we describe shortly. Several mechanisms rapidly terminate the Ca2+ signal and are also responsible for keeping the concentration of Ca2+ in the cytosol low in resting cells. Most importantly, there are Ca2+-pumps in the plasma membrane and the ER membrane that use the energy of ATP hydrolysis to pump Ca2+ out of the cytosol. Cells such as muscle and nerve cells, which make extensive use of Ca2+ signaling, have an additional Ca2+ transporter (a Na+-driven Ca2+ exchanger) in their plasma membrane that couples the efflux of Ca2+ to the influx of Na+.
Feedback Generates Ca2+ Waves and Oscillations
The IP3 receptors and ryanodine receptors of the ER membrane have an important feature: they are both stimulated by low to moderate cytoplasmic Ca2+ concentrations. This Ca2+-induced calcium release (CICR) results in positive feedback, which has a major impact on the properties of the Ca2+ signal. The importance of this feedback is seen clearly in studies with Ca2+-sensitive fluorescent indicators, such as aequorin or fura-2 , which allow researchers to monitor cytosolic Ca2+ in individual cells under a microscope.When cells carrying a Ca2+ indicator are treated with a small amount of an extracellular signal molecule that stimulates IP3 production, tiny bursts of Ca2+ are seen in one or more discrete regions of the cell. These Ca2+ puffs or sparks reflect the local opening of small groups of IP3-gated Ca2+-release channels in the ER. Because various Ca2+-binding proteins act as Ca2+ buffers and restrict the diffusion of Ca2+, the signal often remains localized to the site where the Ca2+ enters the cytosol. If the extracellular signal is sufficiently strong and persistent, however, the local Ca2+ concentration can reach a sufficient level to activate nearby IP3 receptors and ryanodine receptors, resulting in a regenerative wave of Ca2+ release that moves through the cytosol (Figure 15–31), much like an action potential in an axon.
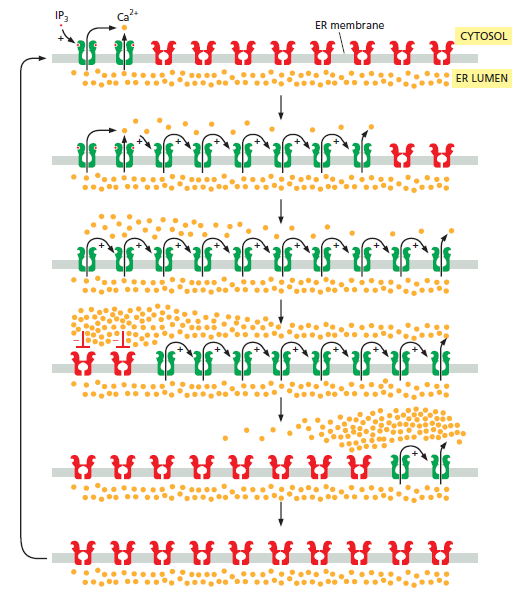
Positive and negative feedback produce Ca2+ waves and oscillations. This diagram shows IP3 receptors and ryanodine receptors on a portion of the ER membrane: active receptors are in green; inactive receptors are in red. When a small amount of cytosolic IP3 activates a cluster of IP3 receptors at one site on the ER membrane (top), the local release of Ca2+ promotes the opening of nearby IP3 and ryanodine receptors, resulting in more Ca2+ release. This positive feedback (indicated by positive signs) produces a regenerative wave of Ca2+ release that spreads across the cell. These waves of Ca2+ release move more quickly across the cell than would be possible by simple diffusion. Also, unlike a diffusing burst of Ca2+ ions, which will become more dilute as it spreads, the regenerative wave produces a high Ca2+ concentration across the entire cell. Eventually, the local Ca2+ concentration inactivates IP3 receptors and ryanodine receptors (middle; indicated by red negative signs), shutting down the Ca2+ release. Ca2+-pumps reduce the local cytosolic Ca2+ concentration to its normal low levels. The result is a Ca2+ spike: positive feedback drives a rapid rise in cytosolic Ca2+, and negative feedback sends it back downagain. The Ca2+ channels remain refractory to further stimulation for some period of time, delaying the generation of another Ca2+ spike (bottom). Eventually, however, the negative feedback wears off, allowing IP3 to trigger another Ca2+ wave. The end result is repeated Ca2+ oscillations . Under some conditions, these oscillations can be seen as repeating narrow narrow waves of Ca2+ moving across the cell.
Another important property of IP3 receptors and ryanodine receptors is that they are inhibited, after some delay, by high Ca2+ concentrations (a form of negative feedback). Thus, the rise in Ca2+ in a stimulated cell leads to inhibition of Ca2+ release; because Ca2+ pumps remove the cytosolic Ca2+, the Ca2+ concentration falls . The decline in Ca2+ eventually relieves the negative feedback, allowing cytosolic Ca2+ to rise again. As in other cases of delayed negative feedback (see Figure 15–18), the result is an oscillation in the Ca2+ concentration. These oscillations persist for as long as receptors are activated at the cell surface, and their frequency reflects the strength of the extracellular stimulus
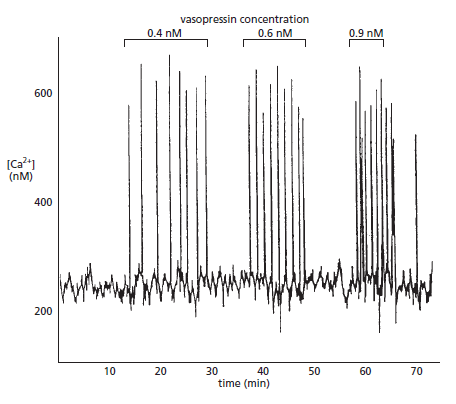
Picture above. Vasopressin-induced Ca2+oscillations in a liver cell. The cell was loaded with the Ca2+-sensitive protein aequorin and then exposed to increasing concentrations of the peptide signal molecule vasopressin, which activates a GPCR and thereby PLCβ . Note that the frequency of the Ca2+ spikes increases with an increasing concentration of vasopressin but that the amplitude of the spikes is not affected. Each spike lasts about 7 seconds.
The frequency, amplitude, and breadth of oscillations can also be modulated by other signaling mechanisms, such as phosphorylation, which influence the Ca2+ sensitivity of Ca2+ channels or affect other components in the signaling system. The frequency of Ca2+ oscillations can be translated into a frequency-dependent cell response. In some cases, the frequency-dependent response itself is also oscillatory: in hormone-secreting pituitary cells, for example, stimulation by an extracellular signal induces repeated Ca2+ spikes, each of which is associated with a burst of hormone secretion. In other cases, the frequency-dependent response is non-oscillatory: in some types of cells, for instance, one frequency of Ca2+ spikes activates the transcription of one set of genes, while a higher frequency activates the transcription of a different set. How do cells sense the frequency of Ca2+ spikes and change their response accordingly? The mechanism presumably depends on Ca2+-sensitive proteins that change their activity as a function of Ca2+-spike frequency.
A protein kinase that acts as a molecular memory device seems to have this remarkable property, as we discuss next.
Ca2+/Calmodulin-Dependent Protein Kinases Mediate Many Responses to Ca2+ Signals
Various Ca2+-binding proteins help to relay the cytosolic Ca2+ signal. The most important is calmodulin, which is found in all eukaryotic cells and can constitute as much as 1% of a cell's total protein mass. Calmodulin functions as a multipurpose intracellular Ca2+ receptor, governing many Ca2+-regulated processes. It consists of a highly conserved, single polypeptide chain with four high-affinity Ca2+-binding sites
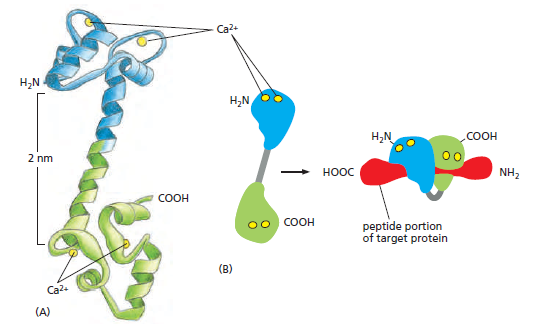
The structure of Ca2+/calmodulin. (A) The molecule has a dumbbell shape, with two globular ends, which can bind to many target proteins. The globular ends are connected by a long, exposed α helix, which allows the protein to adopt a number of different conformations, depending on the target protein it interacts with. Each globular head has two Ca2+- binding sites (Movie 15.6). (B) Shown is the major structural change that occurs in Ca2+/calmodulin when it binds to a target protein (in this example, a peptide that consists of the Ca2+/calmodulin-binding domain of a Ca2+/calmodulin-dependent protein kinase). Note that the Ca2+/calmodulin has “jack-knifed” to surround the peptide. When it binds to other targets, it can adopt different conformations.
When activated by Ca2+ binding, it undergoes a conformational change. Because two or more Ca2+ ions must bind before calmodulin adopts its active conformation, the protein displays a sigmoidal response to increasing concentrations of Ca2+.The allosteric activation of calmodulin by Ca2+ is analogous to the activation of PKA by cyclic AMP, except that Ca2+/calmodulin has no enzymatic activity itself but instead acts by binding to and activating other proteins. In some cases, calmodulin serves as a permanent regulatory subunit of an enzyme complex, but usually the binding of Ca2+ instead enables calmodulin to bind to various target proteins in the cell to alter their activity. When an activated molecule of Ca2+/calmodulin binds to its target protein, the calmodulin further changes its conformation, the nature of which depends on the specific target protein (Figure B above). Among the many targets calmodulin regulates are enzymes and membrane transport proteins. As one example, Ca2+/calmodulin binds to and activates the plasma membrane Ca2+-pump that uses ATP hydrolysis to pump Ca2+ out of cells. Thus, whenever the concentration of Ca2+ in the cytosol rises, the pump is activated, which helps to return the cytosolic Ca2+ level to resting levels. Many effects of Ca2+, however, are more indirect and are mediated by protein phosphorylations catalyzed by a family of protein kinases called Ca2+/calmodulin- dependent kinases (CaM-kinases). Some CaM-kinases phosphorylate transcription regulators, such as the CREB protein, and in this way activate or inhibit the transcription of specific genes. One of the best-studied CaM-kinases is CaM-kinase II, which is found in most animal cells but is especially enriched in the nervous system. It constitutes up to 2% of the total protein mass in some regions of the brain, and it is highly concentrated in synapses. CaM-kinase II has several remarkable properties. To begin
with, it has a spectacular quaternary structure: twelve copies of the enzyme are assembled into a stacked pair of rings, with kinase domains on the outside linked to a central hub
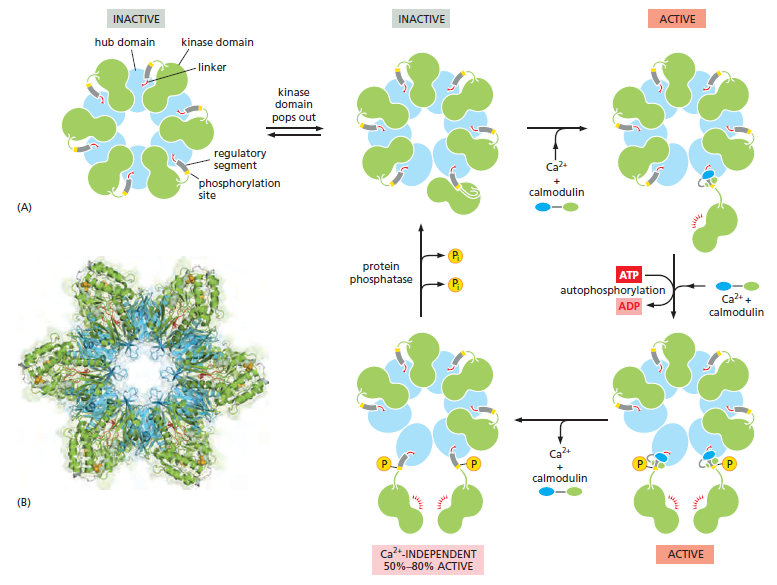
The stepwise activation of CaM-kinase II. (A) Each CaM-kinase II protein has two major domains: an aminoterminal kinase domain (green) and a carboxyl-terminal hub domain (blue), linked by a regulatory segment. Six CaM-kinase II proteins are assembled into a giant ring in which the hub domains interact tightly to produce a central structure that is surrounded by kinase domains. The complete enzyme contains two stacked rings, for a total of 12 kinase proteins, but only one ring is shown here for clarity. When the enzyme is inactive, the ring exists in a dynamic equilibrium between two states. The first (upper left) is a compact state, in which the kinase domains interact with the hub, so that the regulatory segment is buried in the kinase active site and thereby blocks catalytic activity. In the second inactive state (upper middle), a kinase domain has popped out and is linked to the central hub by its regulatory segment, which continues to inhibit the kinase but is now accessible to Ca2+/calmodulin. If present, Ca2+/calmodulin will bind the regulatory segment and prevent it from inhibiting the kinase, thereby locking the kinase in an active state (upper right). If the adjacent kinase subunit also pops out from the hub, it will also be activated by Ca2+/calmodulin, and the two kinases will then phosphorylate each other on their regulatory segments (lower right). This autophosphorylation further activates the enzyme. It also prolongs the activity of the enzyme in two ways. First, it traps the bound Ca2+/calmodulin so that it does not dissociate from the enzyme until cytosolic Ca2+ levels return to basal values for at least 10 seconds (not shown). Second, it converts the enzyme to a Ca2+-independent form, so that the kinase remains active even after the Ca2+/calmodulin dissociates from it (lower left). This activity continues until the action of a protein phosphatase overrides the autophosphorylation activity of CaM-kinase II. (B) This structural model of the enzyme is based on x-ray crystallographic analysis. The remarkable dodecameric structure of the enzyme allows it to achieve a broad range of intermediate activity states in response to different Ca2+ oscillation frequencies: higher frequencies tend to cause more subunits in the enzyme to reach the phosphorylated active state. The behavior of CaM-kinase II is also controlled by the length of the linker segment between the kinase and hub domains. The linker is longer in some isoforms of the enzyme; in these isoforms, the kinase domains tend to pop out of the ring more frequently, making it more sensitive to Ca2+. These and other mechanisms allow the cell to tailor the responsiveness of the enzyme to the needs of different types of neurons.
This structure helps the enzyme function as a molecular memory device, switching to an active state when exposed to Ca2+/ calmodulin and then remaining active even after the Ca2+ signal has decayed. This is because adjacent kinase subunits can phosphorylate each other (a process called autophosphorylation) when Ca2+/calmodulin activates them Once a kinase subunit is autophosphorylated, it remains active even in the absence of Ca2+, thereby prolonging the duration of the kinase activity beyond that of the initial activating Ca2+ signal. The enzyme maintains this activity until a protein phosphatase removes the autophosphorylation and shuts the kinase off. CaM-kinase II activation can thereby serve as a memory trace of a prior Ca2+ pulse, and it seems to have a role in some types of memory and learning in the vertebrate nervous system. Mutant mice that lack a brain-specific form of the enzyme have specific defects in their ability to remember where things are. Another remarkable property of CaM-kinase II is that the enzyme can use its intrinsic memory mechanism to decode the frequency of Ca2+ oscillations. This property is thought to be especially important at a nerve cell synapse, where changes in intracellular Ca2+ levels in a postsynaptic cell as a result of neural activity can lead to long-term changes in the subsequent effectiveness of that synapse When CaM-kinase II is exposed to both a protein phosphatase and repetitive pulses of Ca2+/calmodulin at different frequencies that mimic those observed in stimulated cells, the enzyme’s activity increases steeply as a function of pulse frequency
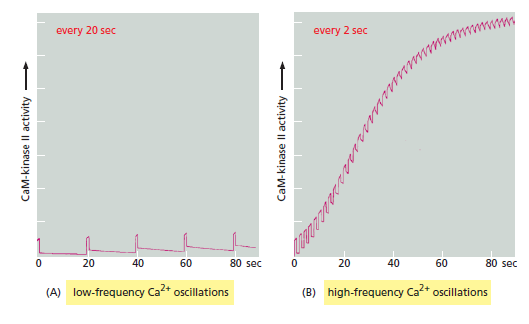
CaM-kinase II as a frequency decoder of Ca2+ oscillations. (A) At low frequencies of Ca2+ spikes, the enzyme becomes inactive after each spike, as the autophosphorylation induced by Ca2+/calmodulin binding does not maintain the enzyme’s activity long enough for the enzyme to remain active until the next Ca2+ spike arrives. (B) At higher spike frequencies, however, the enzyme fails to inactivate completely between Ca2+ spikes, so its activity ratchets up with each spike. If the spike frequency is high enough, this progressive increase in enzyme activity will continue until the enzyme is autophosphorylated on all subunits and is therefore maximally activated. Although not shown, once enough of its subunits are autophosphorylated, the enzyme can be maintained in a highly active state even with a relatively low frequency of Ca2+ spikes (a form of cell memory). The binding of Ca2+/ calmodulin to the enzyme is enhanced by the CaM-kinase II autophosphorylation (an additional form of positive feedback), helping to generate a more switchlike response to repeated Ca2+ spikes.
Some G Proteins Directly Regulate Ion Channels
G proteins do not act exclusively by regulating the activity of membrane-bound enzymes that alter the concentration of cyclic AMP or Ca2+ in the cytosol. The α subunit of one type of G protein (called G12), for example, activates a guanine nucleotide exchange factor (GEF) that activates a monomeric GTPase of the Rho family (discussed later and in Chapter 16), which regulates the actin cytoskeleton. In some other cases, G proteins directly activate or inactivate ion channels in the plasma membrane of the target cell, thereby altering the ion permeability— and hence the electrical excitability—of the membrane. As an example, acetylcholine released by the vagus nerve reduces the heart rate (see Figure 15–5B). This effect is mediated by a special class of acetylcholine receptors that activate the Gi protein discussed earlier. Once activated, the α subunit of Gi inhibits adenylyl cyclase (as described previously), while the βγ subunits bind to K+ channels in the heart muscle cell plasma membrane and open them. The opening of these K+ channels makes it harder to depolarize the cell and thereby contributes to the inhibitory effect of acetylcholine on the heart. (These acetylcholine receptors, which can be activated by the fungal alkaloid muscarine, are called muscarinic acetylcholine receptors to distinguish them from the very different nicotinic acetylcholine receptors, which are ion-channel-coupled receptors on skeletal muscle and nerve cells that can be activated by the binding of nicotine, as well as by acetylcholine.) Other G proteins regulate the activity of ion channels less directly, either by stimulating channel phosphorylation (by PKA, PKC, or CaM-kinase, for example) or by causing the production or destruction of cyclic nucleotides that directly activate or inactivate ion channels. These cyclic-nucleotide-gated ion channels have a crucial role in both smell (olfaction) and vision, as we now discuss.
2. http://creation.com/origin-of-life
Last edited by Otangelo on Sat Mar 02, 2024 3:24 pm; edited 19 times in total