http://reasonandscience.heavenforum.org/t2118-complexity-of-the-cell-s-transport-and-communication-system
Nuclear transport 2
The entry and exit of large molecules from the nucleus is tightly controlled by the nuclear pore complexes. Although small molecules can enter the nucleus without regulation,macromolecules such as RNA and proteins require association karyopherins called importins to enter the nucleus and exportins to exit. "Cargo" proteins that must be translocated from the cytoplasm to the nucleus contain short amino acid sequences known as nuclear localization signals, which are bound by importins, while those transported from the nucleus to the cytoplasm carry nuclear export signals bound by exportins. The ability of importins and exportins to transport their cargo is regulated by GTPases, enzymes that hydrolyze the molecule guanosine triphosphate to release energy. The key GTPase in nuclear transport is Ran, which can bind either GTP or GDP (guanosine diphosphate), depending on whether it is located in the nucleus or the cytoplasm. Whereas importins depend on RanGTP to dissociate from their cargo, exportins require RanGTP in order to bind to their cargo.
Nuclear import depends on the importin binding its cargo in the cytoplasm and carrying it through the nuclear pore into the nucleus. Inside the nucleus, RanGTP acts to separate the cargo from the importin, allowing the importin to exit the nucleus and be reused. Nuclear export is similar, as the exportin binds the cargo inside the nucleus in a process facilitated by RanGTP, exits through the nuclear pore, and separates from its cargo in the cytoplasm. Specialized export proteins exist for translocation of mature mRNA and tRNA to the cytoplasm after post-transcriptional modification is complete. This quality-control mechanism is important due to these molecules' central role in protein translation; mis-expression of a protein due to incomplete excision of exons or mis-incorporation of amino acids could have negative consequences for the cell; thus, incompletely modified RNA that reaches the cytoplasm is degraded rather than used in translation.
Components of Coated Vesicles and Nuclear Pore Complexes Share a Common Molecular Architecture 1
To date three major kinds of transport vesicles, distinguished by the compositions of their protein coat complexes, have been shown to traffic between these internal membranes and the plasma membrane:
First, theclathrin/adaptin complexes are responsible for endocytosis and vesicular trafficking between the Golgi, lysosomes, and endosomes;
second, the COPI complex mediates intra-Golgi and Golgi-to-ER trafficking;
and lastly, the COPII complex supports vesicle movement from the ER to the Golgi
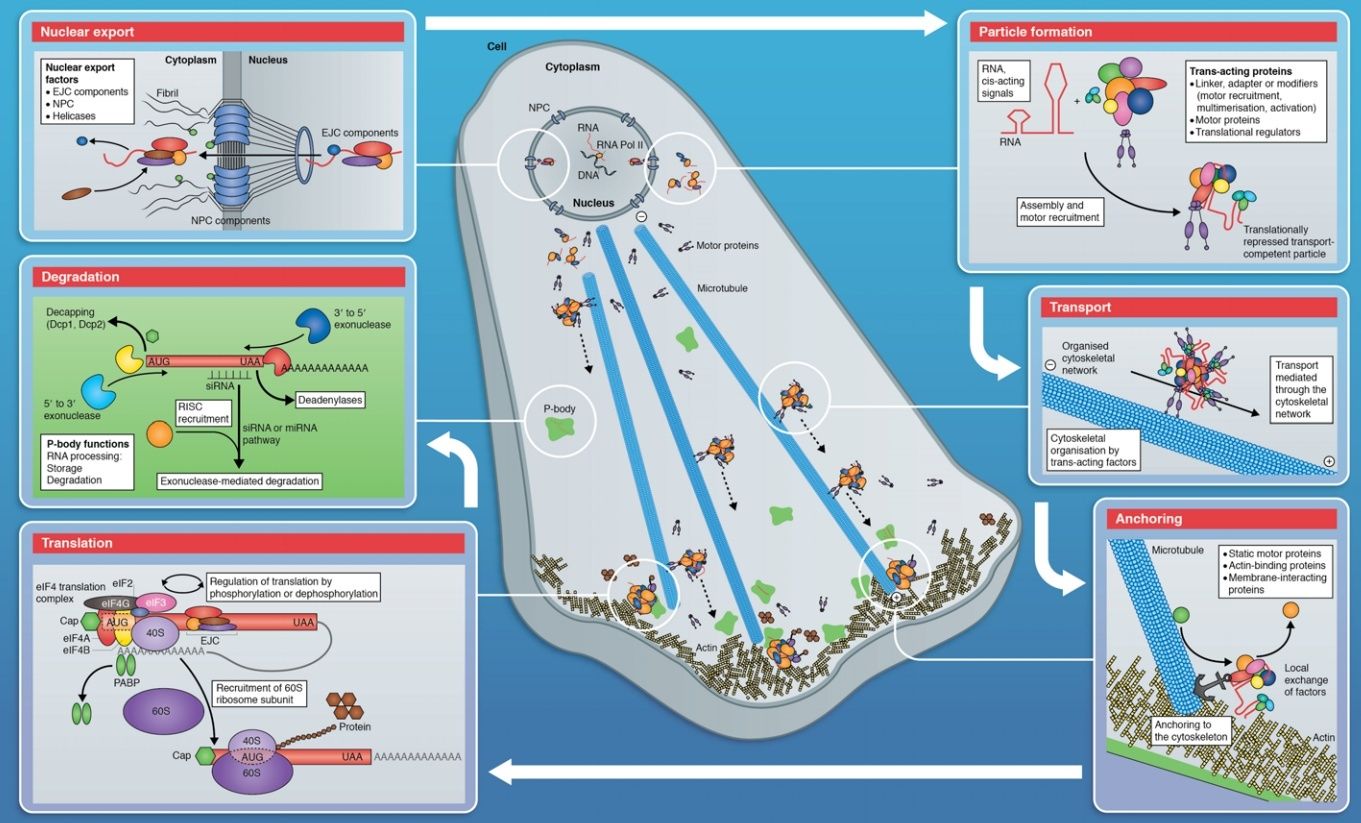
Mature Eukaryotic mRNAs Are Selectively Exported from the Nucleus
Eukaryotic pre-mRNA synthesis and processing take place in an orderly fashion within the cell nucleus. But of the pre-mRNA that is synthesized inside the nucleus of the cell by RNA polymerase, only a small fraction— the mature mRNA—is of further use to the cell. Most of the rest—excised introns, broken RNAs, and aberrantly processed pre-mRNAs—is not only useless but potentially dangerous. How does the cell distinguish between the relatively rare mature mRNA molecules it wishes to keep and the overwhelming amount of debris created by RNA processing? The answer is that, as an RNA molecule is processed, it loses certain proteins and acquires others. For example, we have seen that acquisition of cap-binding complexes, exon junction complexes, and poly-A-binding proteins mark the completion of capping, splicing, and poly-A addition, respectively. A properly completed mRNA molecule is also distinguished by the proteins it lacks. For example, the presence of an snRNP protein would signify incomplete or aberrant splicing. Only when the proteins present on an mRNA molecule collectively signify that processing was successfully completed is the mRNA exported from the nucleus into the cytosol, where it can be translated into protein. Improperly processed mRNAs and other RNA debris (excised intron sequences, for example) are retained in the nucleus, where they are eventually degraded by the nuclear exosome, a large protein complex whose interior is rich in 3ʹ-to-5ʹ RNA exonucleases

.
Eukaryotic cells thus export only useful RNA molecules to the cytoplasm, while debris is disposed of in the nucleus. Of all the proteins that assemble on pre-mRNA molecules as they emerge from transcribing RNA polymerases, the most abundant are the hnRNPs (heterogeneous nuclear ribonuclear proteins). Some of these proteins (there are approximately 30 different ones in humans) unwind the hairpin helices in the RNA so that splicing and other signals on the RNA can be read more easily. Others preferentially package the RNA contained in the very long intron sequences typical in complex organisms (see Figure 6–31) and these may play an important role in distinguishing mature mRNA from the debris left over from RNA processing. Successfully processed mRNAs are guided through the nuclear pore complexes (NPCs)—aqueous channels in the nuclear membrane that directly connect the nucleoplasm and cytosol (Figure 6–37). Small molecules (less than 60,000 daltons) can diffuse freely through these channels. However, most of the macromolecules in cells, including mRNAs complexed with proteins, are far too large to pass through the channels without a special process. The cell uses energy to actively transport such macromolecules in both directions through the nuclear pore complexes. As explained in detail in Chapter 12, macromolecules are moved through nuclear pore complexes by nuclear transport receptors, which, depending on the identity of the macromolecule, escort it from the nucleus to the cytoplasm or vice versa. For mRNA export to occur, a specific nuclear transport receptor must be loaded onto the mRNA, a step that, in many organisms, takes place in concert with 3ʹ cleavage and polyadenylation. Once it helps to move an RNA molecule through the nuclear pore complex, the transport receptor dissociates from the mRNA, re-enters the nucleus, and is then used to export a new mRNA molecule. The export of mRNA–protein complexes from the nucleus can be readily observed with the electron microscope for the unusually abundant mRNA of the insect Balbiani Ring genes. As these genes are transcribed, the newly formed RNA is seen to be packaged by proteins, including hnRNPs, SR proteins, and components of the spliceosome. This protein–RNA complex undergoes a series of structural transitions, probably reflecting RNA processing events, culminating in a curved fiber.

This curved fiber moves through the nucleoplasm and enters the nuclear pore complex (with its 5ʹ cap proceeding first), and it then undergoes another series of structural transitions as it moves through the pore. These and other observations reveal that the pre-mRNA–protein and mRNA–protein complexes are dynamic structures that gain and lose numerous specific proteins during RNA synthesis, processing, and export.
Proteins Can Move Between Compartments in Different Ways
The synthesis of all proteins begins on ribosomes in the cytosol, except for the few that are synthesized on the ribosomes of mitochondria and plastids. Their subsequent fate depends on their amino acid sequence, which can contain sorting signals that direct their delivery to locations outside the cytosol or to organelle surfaces. Some proteins do not have a sorting signal and consequently remain in the cytosol as permanent residents. Many others, however, have specific sorting signals that direct their transport from the cytosol into the nucleus, the ER, mitochondria, plastids, or peroxisomes; sorting signals can also direct the transport of proteins from the ER to other destinations in the cell.
To understand the general principles by which sorting signals operate, it is important to distinguish three fundamentally different ways by which proteins move from one compartment to another. These three mechanisms are described below, and the transport steps at which they operate are outlined in the picture below:
A simplified “roadmap” of protein traffic within a eukaryotic cell. Proteins can move from one compartment to another by
gated transport (red)
protein translocation (blue)
or vesicular transport (green)
The sorting signals that direct a given protein’s movement through the system, and thereby determine its eventual location in the cell, are contained in each protein’s amino acid sequence. The journey begins with the synthesis of a protein on a ribosome in the cytosol and, for many proteins, terminates when the protein reaches its final destination. Other proteins shuttle back and forth between the nucleus and cytosol. At each intermediate station (boxes), a decision is made as to whether the protein is to be retained in that compartment or transported further. A sorting signal may direct either retention in or exit from a compartment. We shall refer to this figure often as a guide in this chapter and the next, highlighting in color the particular pathway being discussed.

The transfer of soluble proteins from the ER to the Golgi apparatus, for example, occurs in this way. Because the transported proteins do not cross a membrane, vesicular transport can move proteins only between compartments that are topologically equivalent

Each mode of protein transfer is usually guided by sorting signals in the transported protein, which are recognized by complementary sorting receptors. If a large protein is to be imported into the nucleus, for example, it must possess a sorting signal that receptor proteins recognize to guide it through the nuclear pore complex. If a protein is to be transferred directly across a membrane, it must possess a sorting signal that the translocator recognizes. Likewise, if a protein is to be loaded into a certain type of vesicle or retained in certain organelles, a complementary receptor in the appropriate membrane must recognize its sorting signal.
Signal Sequences and Sorting Receptors Direct Proteins to the Correct Cell Address
Most protein sorting signals involved in transmembrane transport reside in a stretch of amino acid sequence, typically 15–60 residues long. Such signal sequences are often found at the N-terminus of the polypeptide chain, and in many cases specialized signal peptidases remove the signal sequence from the finished protein once the sorting process is complete. Signal sequences can also be internal stretches of amino acids, which remain part of the protein. Such signals are used in gated transport into the nucleus. Sorting signals can also be composed of multiple internal amino acid sequences that form a specific three-dimensional arrangement of atoms on the protein’s surface; such signal patches are sometimes used for nuclear import and in vesicular transport. Each signal sequence specifies a particular destination in the cell. Proteins destined for initial transfer to the ER usually have a signal sequence at their Nterminus that characteristically includes a sequence composed of about 5–10 hydrophobic amino acids. Many of these proteins will in turn pass from the ER to the Golgi apparatus, but those with a specific signal sequence of four amino acids at their C-terminus are recognized as ER residents and are returned to the ER. Proteins destined for mitochondria have signal sequences of yet another type, in which positively charged amino acids alternate with hydrophobic ones. Finally, many proteins destined for peroxisomes have a signal sequence of three characteristic amino acids at their C-terminus. The table below presents some specific signal sequences.

Question : How could these sequences have evolved ? trial and error ?
Experiments in which the peptide is transferred from one protein to another by genetic engineering techniques have demonstrated the importance of each of these signal sequences for protein targeting. Placing the N-terminal ER signal sequence at the beginning of a cytosolic protein, for example, redirects the protein to the ER; removing or mutating the signal sequence of an ER protein causes its retention in the cytosol. Signal sequences are therefore both necessary and sufficient for protein targeting. Even though their amino acid sequences can vary greatly, the signal sequences of proteins having the same destination are functionally interchangeable; physical properties, such as hydrophobicity, often seem to be more important in the signal- recognition process than the exact amino acid sequence. Signal sequences are recognized by complementary sorting receptors that guide proteins to their appropriate destination, where the receptors unload their cargo. The receptors function catalytically: after completing one round of targeting, they return to their point of origin to be reused. Most sorting receptors recognize classes of proteins rather than an individual protein species. They can therefore be viewed as public transportation systems, dedicated to delivering numerous different components to their correct location in the cell.
The nuclear envelope encloses the DNA and defines the nuclear compartment. This envelope consists of two concentric membranes, which are penetrated by nuclear pore complexes
The nuclear envelope. The double-membrane envelope is penetrated by pores in which nuclear pore complexes (not shown) are positioned. The outer nuclear membrane is continuous with the endoplasmic reticulum (ER). The ribosomes that are normally bound to the cytosolic surface of the ER membrane and outer nuclear membrane are not shown.
The nuclear lamina is a fibrous protein meshwork underlying the inner membrane.

Nuclear Localization Signals Direct Nuclear Proteins to the Nucleus
When proteins are experimentally extracted from the nucleus and reintroduced into the cytosol, even the very large ones reaccumulate efficiently in the nucleus. Sorting signals called nuclear localization signals (NLSs) are responsible for the selectivity of this active nuclear import process. The signals have been precisely defined by using recombinant DNA technology for numerous nuclear proteins, as well as for proteins that enter the nucleus only transiently

In many nuclear proteins, the signals consist of one or two short sequences that are rich in the positively charged amino acids lysine and arginine, with the precise sequence varying for different proteins. Other nuclear proteins contain different signals, some of which are not yet characterized. Nuclear localization signals can be located almost anywhere in the amino acid sequence and are thought to form loops or patches on the protein surface. Many function even when linked as short peptides to lysine side chains on the surface of a cytosolic protein, suggesting that the precise location of the signal within the amino acid sequence of a nuclear protein is not important. Moreover, as long as one of the protein subunits of a multicomponent complex displays a nuclear localization signal, the entire complex will be imported into the nucleus. Macromolecular transport across NPCs differs fundamentally from the transport of proteins across the membranes of other organelles, in that it occurs through a large, expandable, aqueous pore, rather than through a protein transporter spanning one or more lipid bilayers. For this reason, fully folded nuclear proteins can be transported into the nucleus through an NPC, and newly formed ribosomal subunits are transported out of the nucleus as an assembled particle. By contrast, proteins have to be extensively unfolded to be transported into most other organelles.
Nuclear Import Receptors Bind to Both Nuclear Localization Signals and NPC Proteins
To initiate nuclear import, most nuclear localization signals must be recognized by nuclear import receptors, sometimes called importins, most of which are encoded by a family of related genes. Each family member encodes a receptor protein that can bind and transport the subset of cargo proteins containing the appropriate nuclear localization signal

Nuclear import receptors do not always bind to nuclear proteins directly. Additional adaptor proteins can form a bridge between the import receptors and the nuclear localization signals on the proteins to be transported (Figure 1B above). Some adaptor proteins are structurally related to nuclear import receptors. By using a variety of import receptors and adaptors, cells are able to recognize the broad repertoire of nuclear localization signals that are displayed on nuclear proteins. The import receptors are soluble cytosolic proteins that bind both to the nuclear localization signal on the cargo protein and to the phenylalanine-glycine (FG) repeats in the unstructured domains of the channel nucleoporins that line the central pore. FG-repeats are also found in the cytoplasmic and nuclear fibrils. FG-repeats in the unstructured tangle of the pore are thought to do doubleduty. They interact weakly, which gives the protein tangle gel-like properties that impose a permeability barrier to large macromolecules, and they serve as docking sites for nuclear import receptors. FG-repeats line the path through the NPCs taken by the import receptors and their bound cargo proteins. According to one model of nuclear transport, the receptor–cargo complexes move along the transport path by repeatedly binding, dissociating, and then re-binding to adjacent FG-repeat sequences. In this way, the complexes may hop from one nucleoporin to another to traverse the tangled interior of the NPC in a random walk. As import receptors bind to FG-repeats during this journey, they would disrupt interaction between the repeats and locally dissolve the gel phase of the protein tangle that fills the pore, allowing the passage of the receptor–cargo complex. Once inside the nucleus, the import receptors dissociate from their cargo and return to the cytosol. As we will see, this dissociation only occurs on the nuclear side of the NPC and thereby confers directionality to the import process.
Nuclear Export Works Like Nuclear Import, But in Reverse
The nuclear export of large molecules, such as new ribosomal subunits and RNA molecules, occurs through NPCs and also depends on a selective transport system. The transport system relies on nuclear export signals on the macromolecules to be exported, as well as on complementary nuclear export receptors, or exportins. These receptors bind to both the export signal and NPC proteins to guide their cargo through the NPC to the cytosol. Many nuclear export receptors are structurally related to nuclear import receptors, and they are encoded by the same gene family of nuclear transport receptors, or karyopherins. In yeast, there are 14 genes encoding karyopherins; in animal cells, the number is significantly larger. It is often not possible to tell from their amino acid sequence alone whether a particular family member works as a nuclear import or nuclear export receptor. As might be expected, therefore, the import and export transport systems work in similar ways but in opposite directions: the import receptors bind their cargo molecules in the cytosol, release them in the nucleus, and are then exported to the cytosol for reuse, while the export receptors function in the opposite fashion.
In other words: Gate chip code authentication: 4 Here’s a illustration: A round door needs to be open to the environment, but keep interlopers out. Valid users, coming in a wide variety of sizes, need to be allowed access by an automatic authentication system that will usher them in quickly. Once inside, they should not be able to drift back out. The nuclear pore complex appears to use a most elegant solution to this problem of “selective gating.” Imagine a spaceship with a highly-sensitive computer center at its core. Objects and spacemen drift by in this weightless environment. The doors to the computer center must remain open at all times, but entry must be protected from enemies and from those who have no business being in there. Anchored to the rims of these doors are chains that extend outward, drifting about like spaghetti in a breeze tied at one end. The ends of these chains contain crystals that emit a force-field, collectively creating an invisible dome of force around the door, preventing accidental or malicious entry.You, as a valid user, approach the door with a secret crystal in your hand that acts like an authentication chip code. When you extend it toward the chains, they sense it, and rapidly collapse backwards, pulling you in and forming a kind of tunnel around you. The more distant chains are not affected; they continue to stand guard and keep the force field up. Once you are inside, a robotic device removes your code chip and secures it in a protective chamber so that it cannot open the door behind you. Meanwhile, the collapsed chains quickly extend outward again, re-establishing the force field to keep out anything or anybody not having the special chip code.
“The nuclear pore complex regulates cargo transport between the cytoplasm and the nucleus. We set out to correlate the governing biochemical interactions to the nanoscopic responses of the phenylalanineglycine (FG) rich nucleoporin domains, which are involved in attenuating or promoting cargo translocation. We found that binding interactions with the transport receptor karyopherin-[Beta]1 caused the FG domains of the human nucleoporin Nup153 to collapse into compact molecular conformations. This effect was reversed by the action of Ran guanosine triphosphate, which returned the FG domains into a polymer brush-like, entropic barrier conformation. Similar effects were observed in Xenopus oocyte nuclei in situ. Thus, the reversible collapse of the FG domains may play an important role in regulating nucleocytoplasmic transport.”
Question : How could this communication system have evolved ?

Three subsets of sequence complexity and their relevance to biopolymeric information 3
In summary, Sequence complexity can be
1) random (RSC)
2) ordered (OSC)
3) functional (Functional Sequence Complexity)
FSC is the product of nonrandom selection. FSC results from the equivalent of a succession of integrated algorithmic decision node "switch settings." FSC alone instructs sophisticated metabolic function. Self-ordering processes preclude both complexity and sophisticated functions. Self-ordering phenomena are observed daily in accord with chaos theory. But under no known circumstances can self-ordering phenomena like hurricanes, sand piles, crystallization, or fractals produce algorithmic organization. Algorithmic "self-organization" has never been observed despite numerous publications that have misused the term . Bone fide organization always arises from choice contingency, not chance contingency or necessity.
Reduced uncertainty (misnamed "mutual entropy") cannot measure prescriptive information (information that specifically informs or instructs). Any sequence that specifically informs us or prescribes how to achieve success inherently contains choice controls. The constraints of physical dynamics are not choice contingent. Prescriptive sequences are called "instructions" and "programs." They are not merely complex sequences. They are algorithmically complex sequences. They are cybernetic. Random sequences are maximally complex. But they don't do anything useful. Algorithmic instruction is invariably the key to any kind of sophisticated organization such as we observe in any cell. No method yet exists to quantify "prescriptive information" (cybernetic "instructions").
Nucleic acid prescription of function cannot be explained by "order out of chaos" or by "order on the edge of chaos" . Physical phase changes cannot write algorithms. Biopolymeric matrices of high information retention are among the most complex entities known to science. They do not and can not arise from low-informational self-ordering phenomena.
Some proteins involved in the translocation of mRNA through the nuclear pore »
Adaptors as export signals:
Substances, like mRNA, that are transported from the nucleus to the cytoplasm, contain an export signal that serves as a label saying "I should be transported to the cytoplasm". The label is recognized by export receptors. It is probably not the mRNA itself, but rather proteins associated with the mRNA, which are recognized by the export receptors. For example, some common proteins associated with mRNA (like HnRNP A1 and HnRNP K) have been shown to contain export signals.
Also the proteins associated with the 5' end CAP of the mRNA (called Cap Binding Complex CBC) acts as export signal. They are not crucial for the transport of mRNA, but may play a role in the directional translocation since the 5' end of the mRNA is transported in the lead.
Export receptors:
Export receptors bind to the export signals and carry the cargo (the mRNA) to the other side of the nuclear pore complex. In the case of mRNA transport, an export receptor called CRM1 is believed to be important.
Directional transport:
In order for the export protein to bind to the export signal on the cargo, a protein called RanGTP is needed. The whole complex (the mRNA with the proteins acting as export signals, the export receptor and RanGTP) is transported together through the nuclear pore complex, from the nucleus to the cytoplasm. In the cytoplasm, the complex disassociates and in this process RanGTP is hydrolized to RanGDP. The receptors are then recycled to the nucleus.
mRNA may only be transported from the nucleus to the cytoplasm and not in the other direction. The assymetry of RanGTP/RanGDP is thought to be one important factor of this directional transport of mRNA.
1) http://journals.plos.org/plosbiology/article?id=10.1371/journal.pbio.0020380
2) https://en.wikipedia.org/wiki/Cell_nucleus
3) http://www.ncbi.nlm.nih.gov/pmc/articles/PMC1208958/
4) http://crev.info/2007/11/more_cell_codes_and_authentication_mechanisms/#sthash.RvV6qc76.dpuf
5) http://www.nobelprize.org/educational/medicine/dna/a/transport/export_proteins.html
Last edited by Admin on Tue Apr 25, 2017 5:47 pm; edited 31 times in total