https://reasonandscience.catsboard.com/t2070-transfer-rna-and-its-biogenesis
My comment: tRNA's are very specific molecules, and the " made of " follows several steps, requiring a significant number of proteins and enzymes, which are often made of several subunits and ainded by essential co-factors and metals.
The challenge for evolution to the fact, that biological systems incorporate several essential parts, that cannot be eliminated without losing the core function of the system in question, and that these parts have no function of their own and could therefore not be product of natural mechanisms, of gradual evolutionary steps, is in my view more severe than most philosophers of science and scientists like Behe exemplify. In systems of enormous biological complexity like the cell, thousands of parts are essential , many more parts, than the well known examples like the flagellum. Irreducibility is found from the highest level of biological organisation and systems, to a single DNA deoxyribonucleotide, which loses function if reduced to its single components, the bases, phosphate or sugar. Just take off one, and the molecule loses its function. Same goes for the cell. Take off one building block, like the spindle apparatus, and mitosis and cell division is not possible, and life could not reproduce itself.
The make of proteins is similar to the make of cars in a car factory. If the grinder machine to make the motor pistons has a mal function, the pistons cannot be finished, the car's motor block cannot be assembled with all parts, and the motor would not function without that essential part. Amongst thousands of parts, just a tiny one will compromise the function of the whole system. In biological nano-factories, the solutions to overcome problems like damage must all be pre-programmed, and the repair "working horses" to resolve the problem must be ready in place and "know" what to do how, and when. If a roboter in a factory assembly line fails, employees are ready to detect the error and make the repair . In the cell, the mal function of any part even as tiny and irrelevant as it might seem, can be fatal, and if the repair mechanisms are not functioning correctly and fully in place right from the start, the repair can't be done, and life ceases. These repair enzymes which cleave, join, add, replace etc. must be programmed in order to function properly right from the start. Aberrantly processed pre-tRNAs for example are eliminated through a nuclear surveillance pathway by degradation of their 3′ ends, whereas mature tRNAs lacking modifications are degraded from their 5′ends in the cytosol.
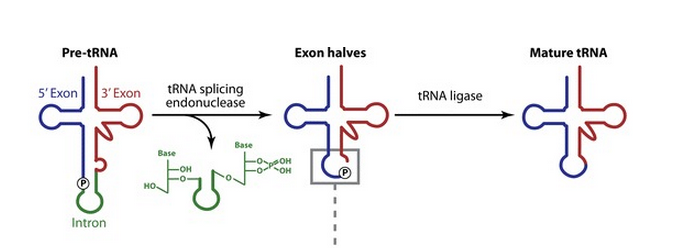
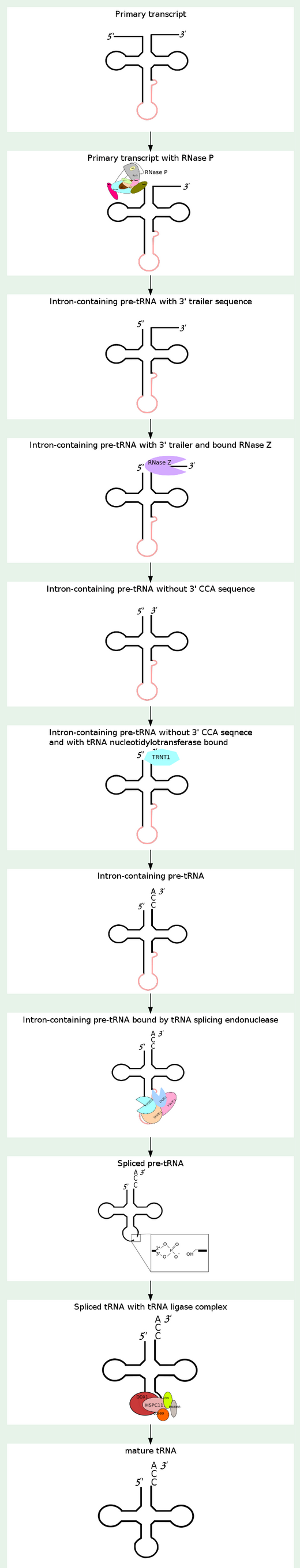
B.Alberts writes: Eucaryotic tRNAs are transcribed from DNA by RNA Polymerase III. Afterwards, tRNA's are covalently modified before they are allowed to exit from the nucleus. Both bacterial and eucaryotic tRNAs are typically synthesized as larger precursor tRNAs, which are then trimmed to produce the mature tRNA. In addition, some tRNA precursors (from both bacteria and eucaryotes) contain introns that must be spliced out. tRNA splicing uses a cut-and-paste mechanism that is catalyzed by proteins. Trimming and splicing both require the precursor tRNA to be correctly folded in its cloverleaf configuration. Because misfolded tRNA precursors will not be processed properly, the trimming and splicing reactions are thought to act as quality- control steps in the generation of tRNA's. All tRNA's are modified chemically—nearly 1 in 10 nucleotides in each mature tRNA molecule is an altered version of a standard G, U, C, or A ribonucleotide. Over 50 different types of tRNA modifications are known. Some of the modified nucleotides—most notably inosine, produced by the deamination of adenosine—affect the conformation and basepairing of the anticodon and thereby facilitate the recognition of the appropriate mRNA codon by the tRNA molecule. This means, if the basepairing of the codons of mRNA with the anticodons of tRNA does not fit and match correctly,it will affect the accuracy with which the correct amino acid is attached to the tRNA , or it is eventually not even capable of identifiyng the right tRNA. In other words, its like the key that must fit in the door lock. It it does not fit, the door will not open. If the match of the codons do not fit precisely into the anticodon's of the mRNA, the precise assignment of the amino acid is compromised, or not possible, and proteic amino acid chains cannot be sinthesized successfully. So that is another keystep.
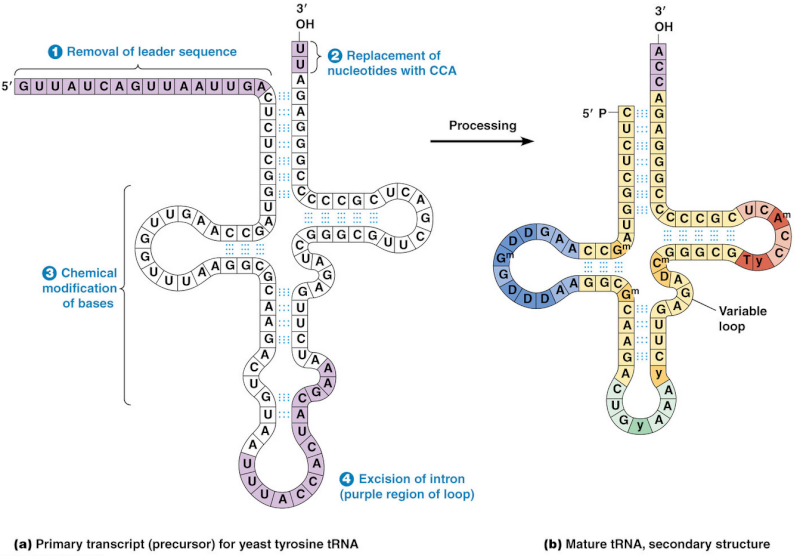
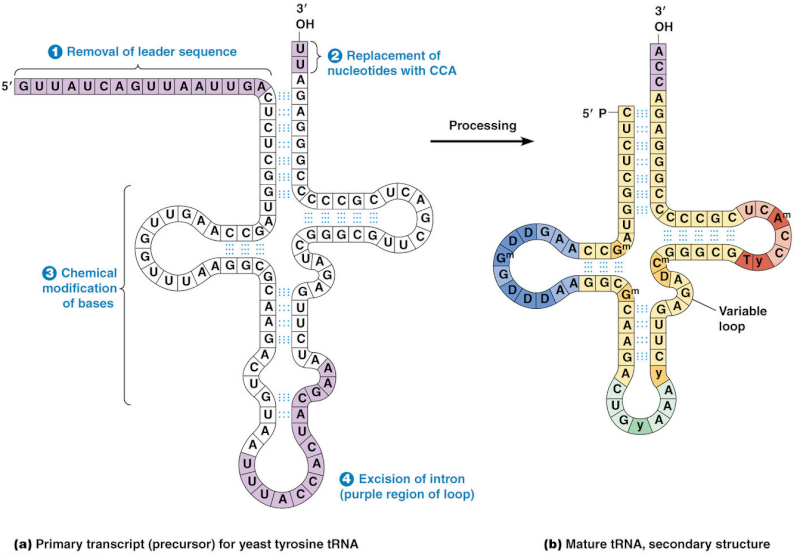
The processing into mature tRNA happens through the removal, addition and chemical modification of nucleotides. Processing for some tRNA involves
1) removal of the leader sequence at the 5 prime end
2) replacement of two nucleotides at the 3 prime end by the sequence CCA (with which all mature tRNA molecules terminate)
3) chemical modification of certain bases and
4) excision of introns. The mature tRNA is often diagrammed as a flattened cloverleaf which clearly shows the base pairing between self-complementary stretches in the molecule.
Each of these steps is a essential requirement for the synthesis of tRNA, if one doesn't do its job properly, tRNA cannot be made. The biosynthesis of tRNA is a irreducible complex process.
To give a example in tRNA maturation in Homo sapiens, following Enzymatic complexes are involved in the process:
Proteins:
CCA tRNA nucleotidyltransferase 1 ,
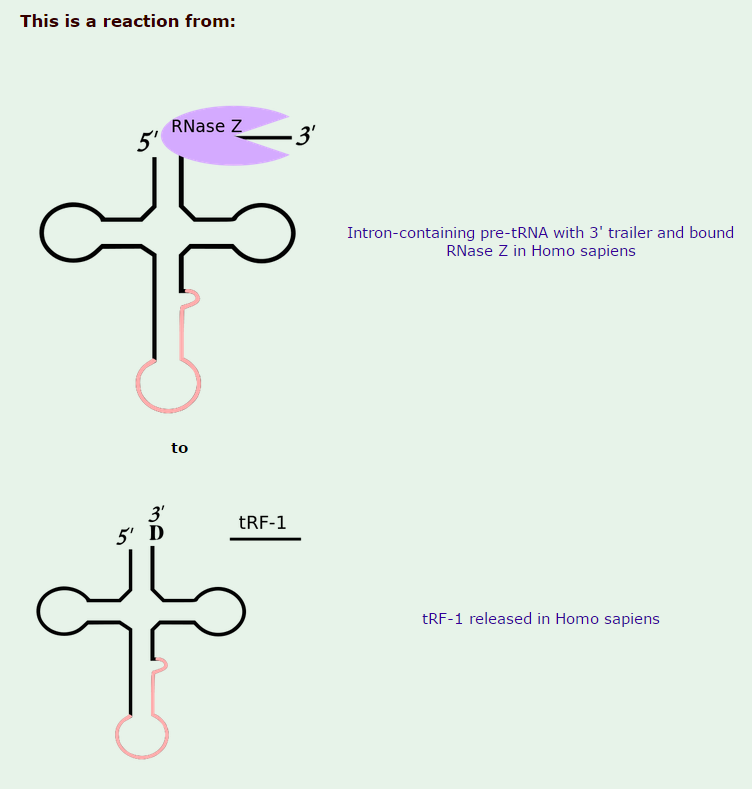
Zinc phosphodiesterase ELAC protein 2
and Enzymatic complexes:
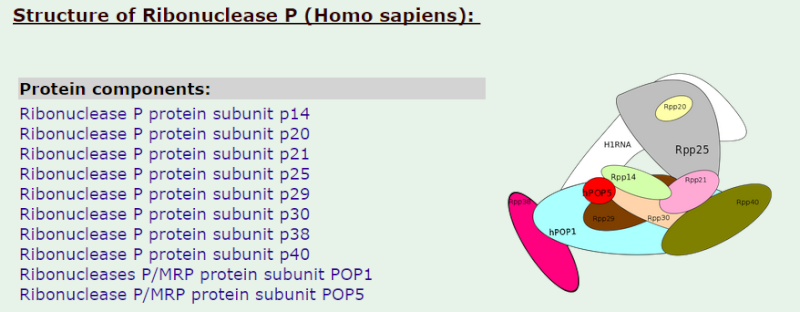
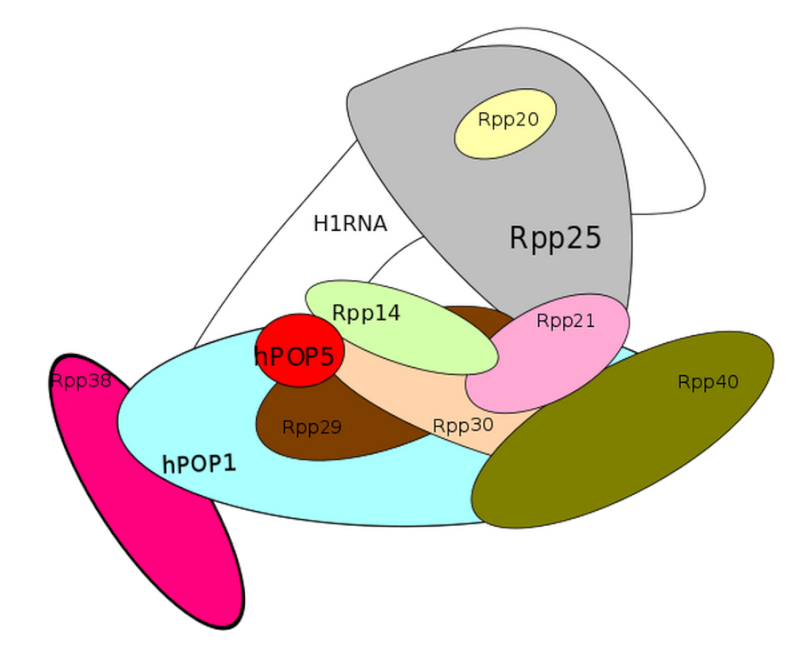
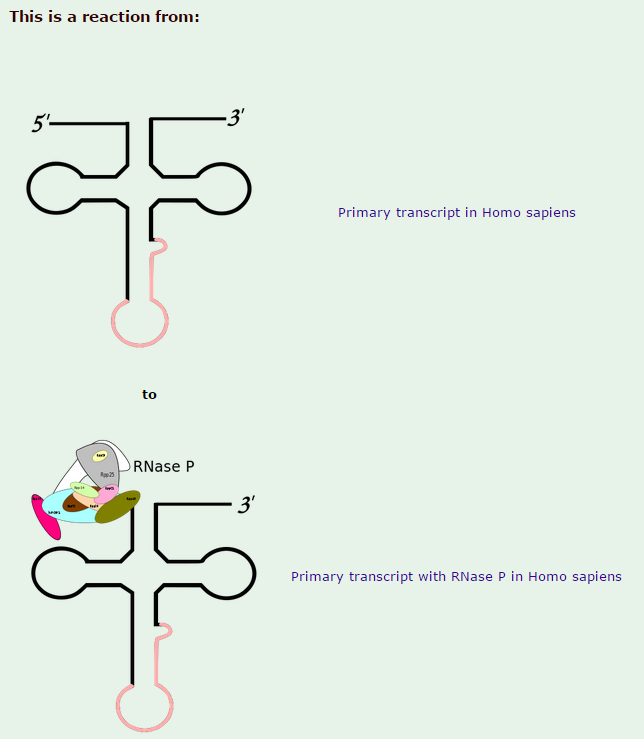
Ribonuclease P
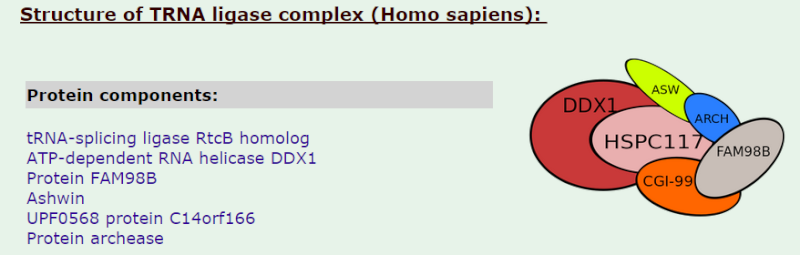
tRNA ligase complex
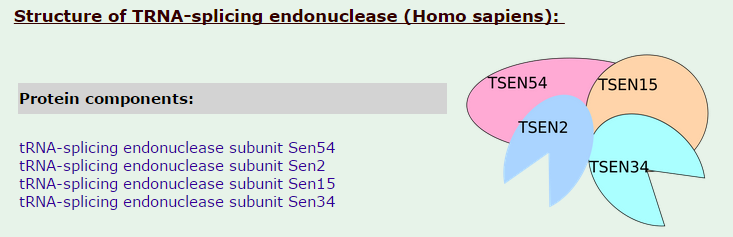
tRNA-splicing endonuclease
CCA tRNA nucleotidyltransferase 1 uses a Magnesium co-factor, Zinc phosphodiesterase ELAC protein 2 uses zinc as co-factor, Human nuclear RNase P consists of 10 Protein subunits and one RNA subunit, the tRNA ligase complex uses 6 protein components, and tRNA-splicing endonuclease uses 4 protein subunits. In total 20 proteins subunits, one RNA subunit, and 2 different co-factors.
Each of these protein complexes exercises very precise coordinated tasks, which all have to be pre-programmed in the genome. Lets have a look at the special capabilities:
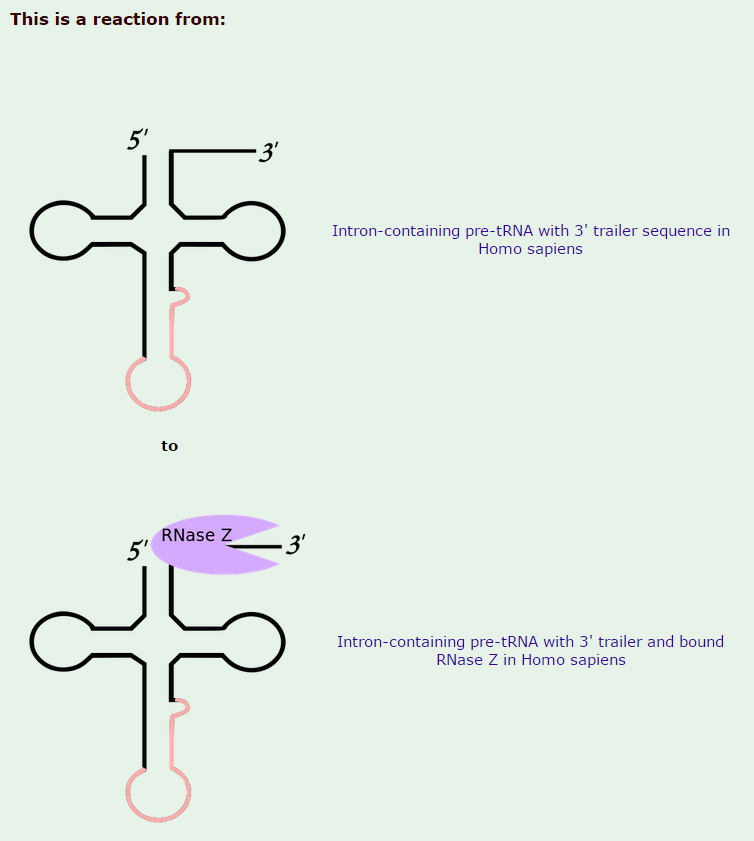
Ribonuclease P has the function to cleave off an extra, or precursor, sequence of RNA on tRNA molecules. For ( supposedly ) billions of years and still to this day, the function of RNase P -- found in nearly all organisms, from bacteria to humans -- has been to cleave transfer tRNA. If the tRNA is not cleaved, it is not useful to the cell.
Once RNase P recognizes tRNA, it docks and, assisted by metal ions, cuts one chemical bond.
My comment: This happens in a stepwise, orderly process, where the enzyme " knows " exactly where to cleave with a precise target. How could such a function have arisen ? trial and error ? coding the genetic instructions until the right sequence permitted to cleave off the right nucleotides ? why at all would some unknown mechanism do this trial and error ? Or had chemicals a end goal ? or the goal of " survival of the fittest " ( despite the fact that they are not alive ) ? if the enzyme cleaved too much or too less, tRNA could not be used properly, so its function had to be programmed correctly in the genome right from the start, otherwise, well, no life.... Not only the cleavage at the right place has to be explained, but also the arise of this sophisticated mechanism, which follows precise , complex steps in a machinelike manner.
In the paper The enigma of ribonuclease P evolution, the authors, Enno Roland K. Hartmann write :
The simplest interpretation is that RNase P has an ‘RNA-alone’ origin and progenitors of Bacteria and Archaea diverged very early in evolution and then pursued completely different strategies in the recruitment of protein subunits during the transition from the ‘RNA-alone’ to the ‘RNA-protein’ state of the enzyme.’
My comment: The authors write about recruitment and strategies. Its interesting that they atribute mental and conscient activities to chemical processes and reactions. But as such, they have no end goal, so how does it make sense to write in these terms ? Furthermore, recruitment of what ? of extant subunits ? were they readily available to choose from in the surrounding ? how could RNase know which ones to select and how to incorporate them correctly in its system ? Is that not one more nice example of pseudo science ?
As Luskin of the discovery institute writes : When certain biologists discuss the early stages of life there is a tendency to think too vaguely. They see a biological wonder before them and they tell a story about how it might have come to be. They may even draw a picture to explain what they mean. Indeed, the story seems plausible enough, until you zoom in to look at the details. I don't mean to demean the intelligence of these biologists. It's just that it appears they haven't considered things as completely as they should. Like a cartoon drawing, the basic idea is portrayed, but there is nothing but blank space where the profound detail of biological processes should be.
Would these five conditions not have to be met in order to recruit and insert the subunits into the system ?
C1: Availability. Among the parts available for recruitment to form the system, there would need to be ones capable of performing the highly specialized tasks of individual parts, even though all of these items serve some other function or no function.
C2: Synchronization. The availability of these parts would have to be synchronized so that at some point, either individually or in combination, they are all available at the same time.
C3: Localization. The selected parts must all be made available at the same ‘construction site,’ perhaps not simultaneously but certainly at the time they are needed.
C4: Coordination. The parts must be coordinated in just the right way: even if all of the parts of a system are available at the right time, it is clear that the majority of ways of assembling them will be non-functional or irrelevant.
C5: Interface compatibility. The parts must be mutually compatible, that is, ‘well-matched’ and capable of properly ‘interacting’: even if sub systems or parts are put together in the right order, they also need to interface correctly.
( Agents Under Fire: Materialism and the Rationality of Science, pgs. 104-105 (Rowman & Littlefield, 2004). HT: ENV.)
In the paper tRNA-nucleotidyltransferases: Highly unusual RNA polymerases with vital functions, the authors Stefan Vörtler, and Mario Mörl write:
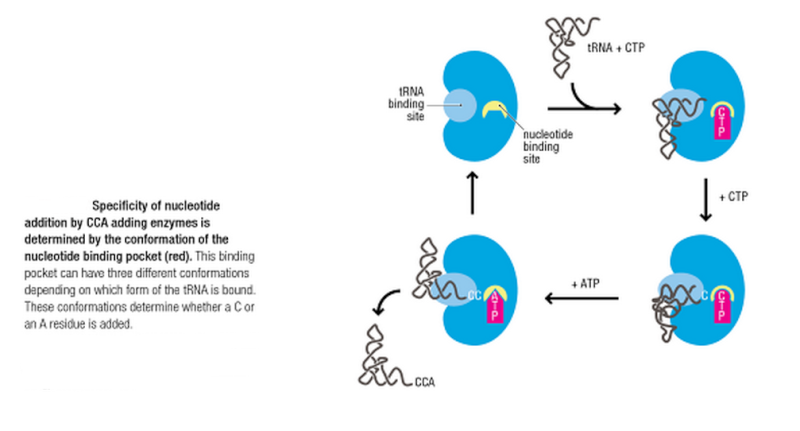
tRNA-nucleotidyltransferases are fascinating and unusual RNA polymerases responsible for the synthesis of the nucleotide triplet CCA at the 3′-terminus of tRNAs. As this CCA end represents an essential functional element for aminoacylation and translation, these polymerases (CCA-adding enzymes) are of vital importance in all organisms. Elucidation of the role of the CCA enzyme in the cellular network of tRNA quality control and the identities of the RNases accompanying the CCA enzyme constitute new questions that warrant active investigation.
CCA-adding enzymes obviously can count until three: after the addition of three nucleotides, the polymerization reaction is efficiently stopped. Additionally, and most interestingly, the CCA-adding enzymes recognize if nucleotides are previously added to a tRNA primer and incorporate then only the missing ones, completing thereby the CCA triplet. A tRNA that carries already the first C residue of the CCA terminus is elongated only by one C and one A, while on a tRNA ending with CC, only the terminal A residue is added. This feature shows that CCA-adding enzymes are not only responsible for the de novo synthesis of CCA ends but have an important maintenance and repair function for tRNA ends. This stringent sequence and length control of the tRNA CCA end reflects the recognition requirements for aminoacylation and translation.
( This is amazing. How did it " learn " that feat ? trial and error ? )
Furthermore, positioning in the ribosome during translation and even peptide release from the ribosome depend on an intact CCA end, which is critical for water coordination and efficient hydrolysis of the ester bound translation product.
These facts indicate that an accurate CCA end participates, beyond simple recognition and binding, as an integral part in several reaction mechanisms and is therefore of vital importance for the cell.
Surprisingly, these polymerases with such unusual features evolved twice in evolution, leading to classes 1 and 2 CCA-adding enzymes
Convergence is evidence against evolution, and the author supposes evolution prior the existence of a replicating cell.......
While class 1 is exclusively found in archaea, class 2 tRNA-nucleotidyltransferases are present in eukaryotes and bacteria, where they fulfill identical functions. Structural organization of classes 1 and 2 CCA-adding enzymes. While both enzyme versions have a hook-like shape of similar size, the allocation of secondary structure elements in neck, body and tail domains are quite different. In class 1 enzymes, these regions contain alpha-helical as well as beta-sheet elements. Class 2, on the other hand, has exclusively alpha-helical structures in these domains. The catalytic cores, located in head and neck domains of both enzyme versions, are indicated by the grey arrows. The rainbow color bar represents the consecutive protein regions from N- (blue) to C-terminus (red).
One of the most fascinating aspects of both classes of tRNA-nucleotidyltransferases is the fact that CCA-addition does not require an external nucleic acid as a template – somehow these enzymes “know” when to incorporate which nucleotide.
Indeed. Or maybe the intelligent designer programmed them in order for them to know ?? what makes more sense, inanimated matter to know something, or a intelligent creator programming these enzymes to exercise special tasks and functions upon pre-programming ?
Crystal structures of both classes 1 and 2 enzymes revealed a set of highly conserved amino acid residues located in the single nucleotide binding pocket that interact with the incoming nucleotide by forming Watson/Crick-like hydrogen bonds.
So these enzymes do not only " know " when to incorporate which nucleotide, but also " know " how to bind each nucleotide to the next through hydrogen bonds..... amazing.
Structural organization of classes 1 and 2 CCA-adding enzymes. While both enzyme versions have a hook-like shape of similar size, the allocation of secondary structure elements in neck, body and tail domains are quite different. In class 1 enzymes, these regions contain alpha-helical as well as beta-sheet elements. Class 2, on the other hand, has exclusively alpha-helical structures in these domains. The catalytic cores, located in head and neck domains of both enzyme versions, are indicated by the grey arrows. The rainbow color bar represents the consecutive protein regions from N- (blue) to C-terminus (red).
One of the most fascinating aspects of both classes of tRNA-nucleotidyltransferases is the fact that CCA-addition does not require an external nucleic acid as a template – somehow these enzymes “know” when to incorporate which nucleotide.
Indeed. Isn't that a magnificient example and evidence of design ?
Crystal structures of both classes 1 and 2 enzymes revealed a set of highly conserved amino acid residues located in the single nucleotide binding pocket that interact with the incoming nucleotide by forming Watson/Crick-like hydrogen bonds
My comment: So these enzymes do not only " know " when to incorporate which nucleotide, but also " know " how to bind each nucleotide to the next through hydrogen bonds..... amazing.
So the question arises : Did natural processes have foresight of the end product, tRNA, to make these highly specific nano robot - like molecular machines which remove, add and modify the nucleotides of tRNA? If not, how could they have arisen, since without end goal, there would be no function for them ? neither could they have been co-opted because of their high specificity and uniqueness, required only in these molecular machines? They are specifically made for the production and make of tRNA's. Isnt the make of tRNA not another prime example of intelligent design ? My contemption is once more that naturalistic explanations are inadequate to explain this sophisticated mechanism in question. While a designer, which had the intention to make life, could have well invented the process, and set it up.
Transfer RNA is an ancient molecule, central to every task a cell performs and thus essential to all life.
The enzyme is one of only two ribozymes which can be found in all kingdoms of life (Bacteria, Archaea, and Eukarya). 11
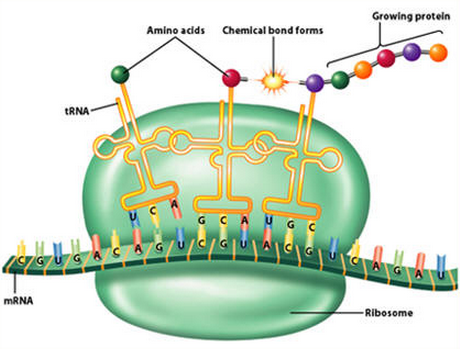
The three major RNAs involved in the flow of genetic information are messenger RNA (mRNA), ribosomal RNA (rRNA), and transfer RNA (tRNA). All these RNAs participate in the protein-synthesizing pathway in cells. tRNA has two distinct characteristics. It carries an anticodon corresponding to the mRNA codon and it binds to the corresponding amino acid in a reaction catalyzed by a specific aminoacyl-tRNA synthetase. In this sense, tRNA is a key bridging molecule between ribonucleotide information (RNA world) and peptide information (protein world). Therefore, tracing the origin of tRNA molecules is likely to cast light on the processes that led to the establishment of the central dogma. 16
10
Of the thousands of RNAs so far identified, transfer RNA (tRNA) is the most direct intermediary between genes and proteins. Like many other RNAs (ribonucleic acids), tRNA aids in translating genes into the chains of amino acids that make up proteins. With the help of a highly targeted enzyme, each tRNA molecule recognizes and latches onto a specific amino acid, which it carries into the protein-building machinery. In order to successfully add its amino acid to the end of a growing protein, tRNA must also accurately read a coded segment of messenger RNA, which gives instructions for the exact sequence of amino acids in the protein. see here
tRNAs Are covalently modified before they exit from the Nucleus
Like most other eucaryotic RNAs, tRNAs are covalently modified before they are allowed to exit from the nucleus.Eucaryotic tRNAs are synthesized by RNA Polymerase III. Both bacterial and eucaryotic tRNAs are typically synthesized as larger precursor tRNAs, which are then trimmed to produce the mature tRNA. In addition, some tRNA precursors (from both bacteria and eucaryotes) contain introns that must be spliced out. This splicing reaction differs chemically from pre-mRNA splicing; rather than generating a lariat intermediate, tRNA splicing uses a cut-and-paste mechanism that is catalyzed by proteins See below:
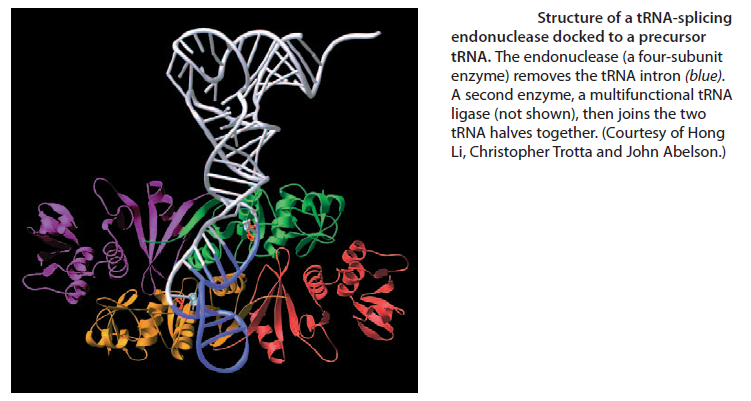
Trimming and splicing both require the precursor tRNA to be correctly folded in its cloverleaf configuration. Because misfolded tRNA precursors will not be processed properly, the trimming and splicing reactions are thought to act as quality- control steps in the generation of tRNAs. All tRNAs are modified chemically—nearly 1 in 10 nucleotides in each mature tRNA molecule is an altered version of a standard G, U, C, or A ribonucleotide.Over 50 different types of tRNA modifications are known; a few are shown below:
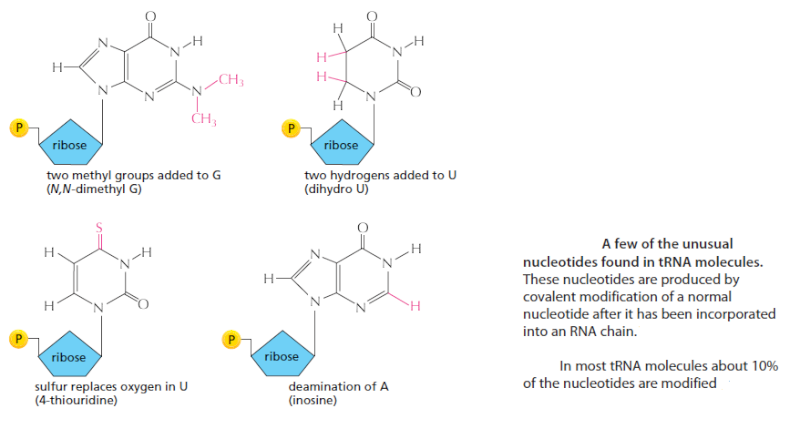
Some of the modified nucleotides—most notably inosine, produced by the deamination of adenosine—affect the conformation and basepairing of the anticodon and thereby facilitate the recognition of the appropriate mRNA codon by the tRNA molecule. Others affect the accuracy with which the tRNA is attached to the correct amino acid.Some of the modified nucleotides—most notably inosine, produced by the deamination of adenosine—affect the conformation and basepairing of the anticodon and thereby facilitate the recognition of the appropriate mRNA codon by the tRNA molecule . Others affect the accuracy with which the tRNA is attached to the correct amino acid.[/b]
After tRNA is transcribed by RNA polymerase III as a precursor tRNA, it must be processed into a mature tRNA
This happens through the removal, addition and chemical modification of nucleotides. Processing for some tRNA involves 2
1) removal of the leader sequence at the 5 prime end
2) replacement of two nucleotides at the 3 prime end by the sequence CCA (with which all mature tRNA molecules terminate)
3) chemical modification of certain bases and
4) excision of an intron.
The mature tRNA is often diagrammed as a flattened cloverleaf which clearly shows the base pairing between self-complementary stretches in the molecule.
tRNA maturation in Homo sapiens
Enzymatic complexes involved in the process: 18
Proteins:
CCA tRNA nucleotidyltransferase 1
Zinc phosphodiesterase ELAC protein 2
Enzymatic complexes:
Ribonuclease P
tRNA ligase complex
tRNA-splicing endonuclease
17
19
 16
16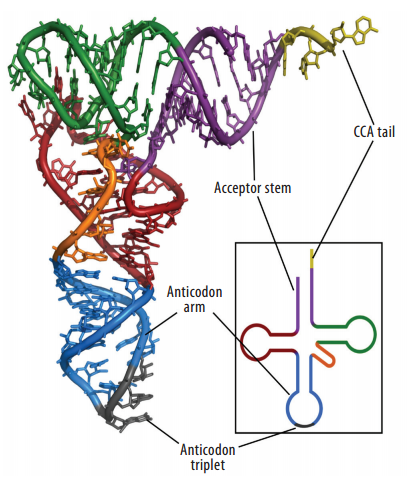
1.The transcription product, the pre-tRNA, contains additional RNA sequences at both the 5’ and 3’-ends. These additional sequences are removed from the transcript during processing. The additional nucleotides at the 5’-end are removed by an unusual RNA containing enzyme called ribonuclease P (RNase P)
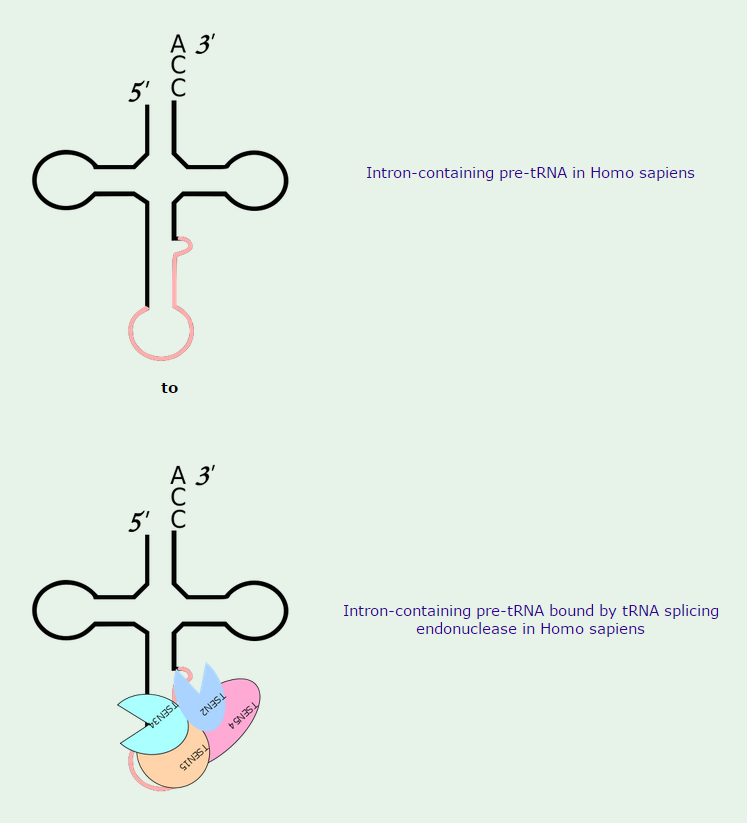
2.Some tRNA precursors contain an intron located in the anticodon arm. These introns are spliced out during processing of the tRNA.
The cloverleaf structure of a single polynucleotide tRNA molecule is universally conserved among organisms. However, tRNA genes are often divided into parts on the chromosome; in bacteria, archaea, eukarya, and organelles, several tRNA genes are interrupted by various types of introns, which are removed by RNA splicing after transcription
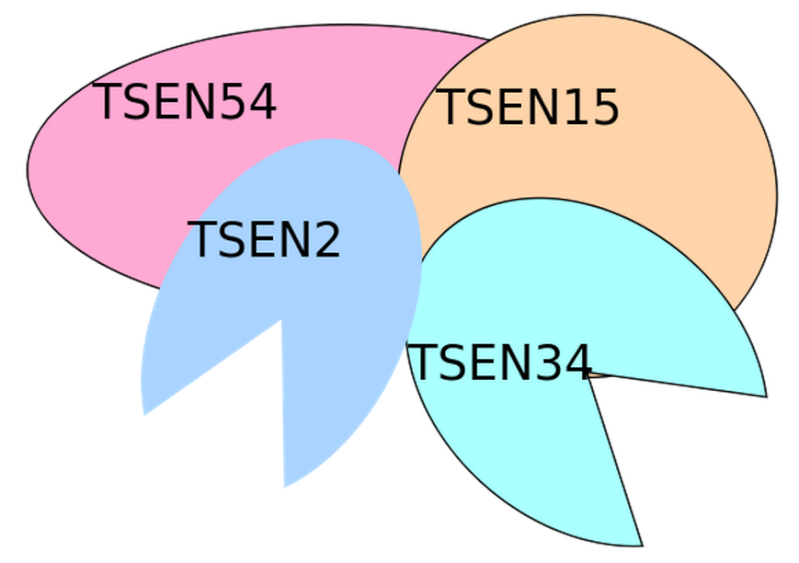
Introns in nuclear and archaeal tRNAs are generally cleaved by tRNA-splicing endonuclease 13
tRNA splicing is a fundamental process required for cell growth and division. SEN2 is a subunit of the tRNA splicing endonuclease, which catalyzes the removal of introns, the first step in tRNA splicing 14
The tRNA splicing reaction in yeast occurs in three steps; each step is catalyzed by a distinct enzyme, which can function interchangeably on all of the substrates 15
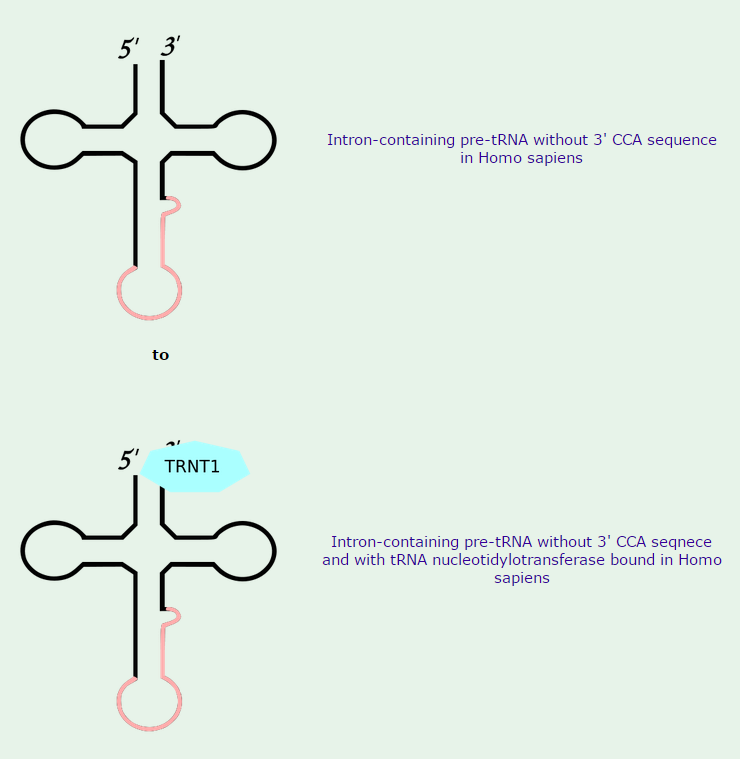
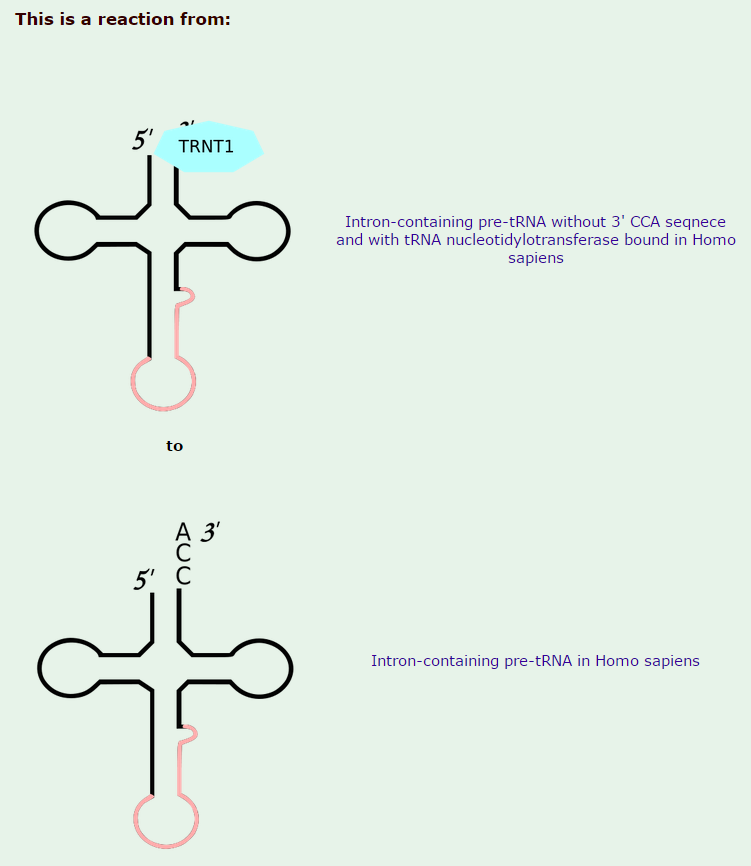
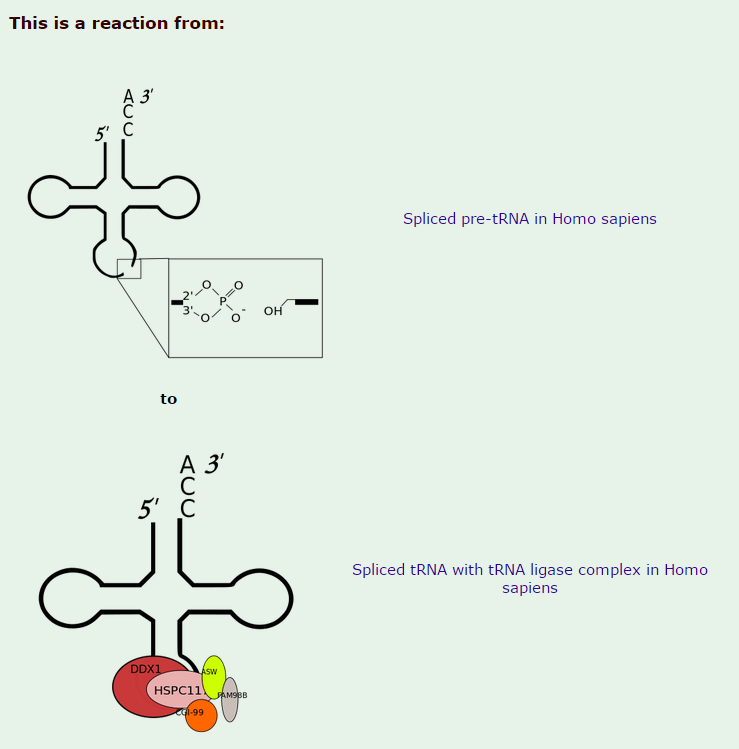
3.All mature tRNAs contain the trinucleotide CCA at their 3’-end. These three bases are not coded for by the tRNA gene. Instead, these nucleotides are added during processing of the pre-tRNA transcript. The enzyme responsible for the addition of the CCA-end is tRNA nucleotidyl transferase and the reaction proceeds according to the following scheme:
tRNA +CTP --> tRNA-C + PPi (pyrophosphate)
tRNA-C +CTP --> tRNA-C-C + PPi
tRNA-C-C +ATP --> tRNA-C-C-A + PPi
4.Mature tRNAs can contain up to 10% bases other than the usual adenine (A), guanine (G), cytidine (C) and uracil (U). These base modifications are introduced into the tRNA at the final processing step. The biological function of most of the modified bases is uncertain and the translation process seems normal in mutants lacking the enzymes responsible for modifying the bases.
Termination signals end the transcription of RNA by RNA polymerase I and RNA polymerase III without the activity of hairpin structures as seen in prokaryotes.
mRNA is cleaved 10 to 35 base-pairs downstream of a AAUAAA sequence (which acts as a poly-A tail addition signal).
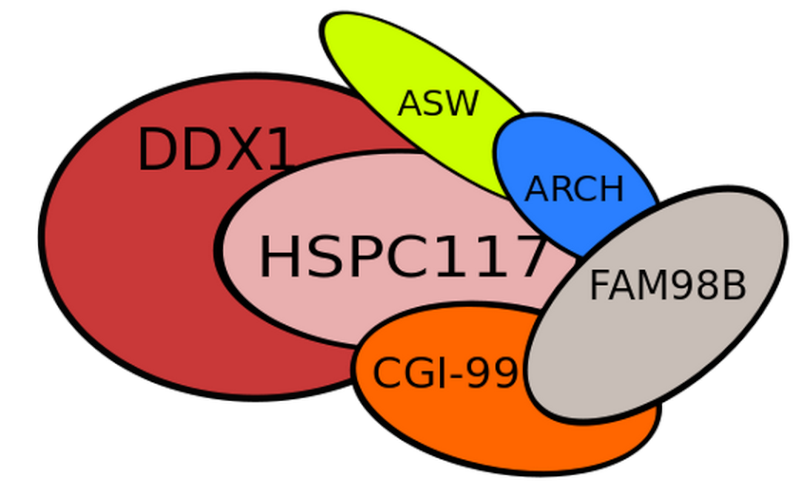
The biogenesis of mature transfer (t)RNAs in cells has a complexity that belies the elegance of their function as the adaptor molecules of protein synthesis. They are synthesized as precursors which are converted to mature tRNA molecules by a sequence of events that includes processing of their 5′ and 3′ ends, modification of a number of bases, addition of the terminal CCA residues and, in the case of intron-containing tRNAs, splicing 1
Transfer RNAs (tRNAs) play an important role linking mRNA and amino acids during protein biogenesis. Four types of tRNA genes have been identified in living organisms. However, the evolutionary origin of tRNAs remains largely unknown. Given their central role in life and their high level of conservation, tRNAs have stimulated extensive interest in their origin and evolution
My comment: This enzyme alone is like a roboter in a assembly line in a factory. If you take it away, the whole mechanism of protein synthesis ceases to exist. No RNase, no proteinsynthesis, no life....... As with RNase, the Protein production needs hundreds, well, probably thousands of intricate fine tuned parts, all doing their function in a precise way, and if you take away just one small apparently irrelevant part, bye bye proteins, bye bye life. These enzyme subparts have no use by their own, but only if correctly inserted in the intricate holoenzyme complexes exercising precisely their function.........
further readings :
http://journal.frontiersin.org/researchtopic/1729/molecular-biology-of-the-transfer-rna-revisited
Molecular biology of the transfer RNA revisited
Last edited by Otangelo on Fri Jan 01, 2021 5:26 am; edited 35 times in total



 ´
´