https://reasonandscience.catsboard.com/t2058-transfer-rna-and-its-biogenesis
tRNA's are very specific and complex molecules, and the " made of " follows several steps, requiring a significant number of proteins and enzymes, which are by themselves also enormously complex, not only in their structure but as well in their " made of ". So the question in the end arises: did natural processes have the foresight of the end product, tRNA, to make this highly specific nanorobot - like molecular machines which remove, add and modify the nucleotides? If not, how could they have arisen, since, without end goal, there would be no function for them? these enzymes are all specifically made for the production of tRNAs. And tRNA is essential for life
JOSEF BERGER: THE GENETIC CODE AND THE ORIGIN OF LIFE 1976
If the t-RNA's of recent organisms have about 80 nucleotides, then the random origin of recent forms of tRNA's has a low probability because that of the origin of one definite t-RNA molecule is about 1: 10^54. The probability of the origin of one definite protein molecule is also slight, i.e. 1: 10^130 if we take into consideration small protein subunits with 100 amino acids. The probability of the random independent simultaneous origin of one definite protein
molecule and one definite nucleic acid molecule is even 1: 10^184 in our case. These and others facts, taking into consideration that the age of the earth is 10^17 seconds. only, can be a basis for hypotheses on an extra-terrestrial origin of life. On the other hand Portelli (1975) defends that 'the complex and perfect information existing in the genetic code is (explained by) that the evolutionary cycles of the universe and the cycle of life are interconnected, namely the information accumulated by the development of life, within the framework of a cycle of the universe passes through the origin of the next universe in the new cosmic cycle. The actual genetic code represents just the informational message arising from the living systems of the previous universe'. However, we have not a single fact as indication for the existence of such a process.
Transfer RNA is an ancient molecule, central to every task a cell performs and thus essential to all life.
The enzyme is one of only two ribozymes which can be found in all kingdoms of life (Bacteria, Archaea, and Eukarya). 11
The three major RNAs involved in the flow of genetic information are messenger RNA (mRNA), ribosomal RNA (rRNA), and transfer RNA (tRNA). All these RNAs participate in the protein-synthesizing pathway in cells. tRNA has two distinct characteristics. It carries an anticodon corresponding to the mRNA codon and it binds to the corresponding amino acid in a reaction catalyzed by a specific aminoacyl-tRNA synthetase. In this sense, tRNA is a key bridging molecule between ribonucleotide information (RNA world) and peptide information (protein world). Therefore, tracing the
10
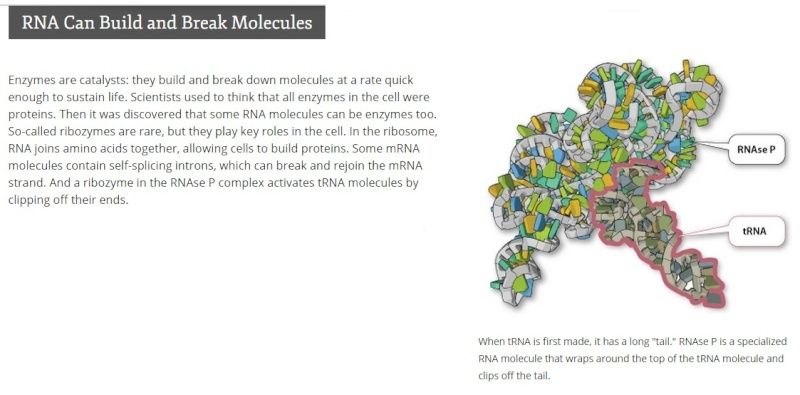
Of the thousands of RNAs so far identified, transfer RNA (tRNA) is the most direct intermediary between genes and proteins. Like many other RNAs (ribonucleic acids), tRNA aids in translating genes into the chains of amino acids that make up proteins. With the help of a highly targeted enzyme, each tRNA molecule recognizes and latches onto a specific amino acid, which it carries into the protein-building machinery. In order to successfully add its amino acid to the end of a growing protein, tRNA must also accurately read a coded segment of messenger RNA, which gives instructions for the exact sequence of amino acids in the protein. see here
tRNAs Are covalently modified before they exit from the Nucleus
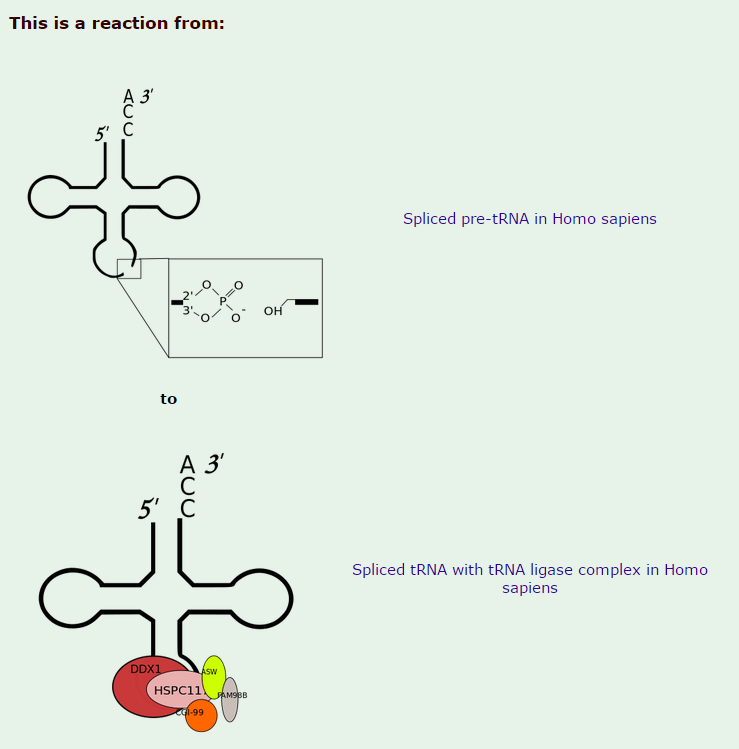
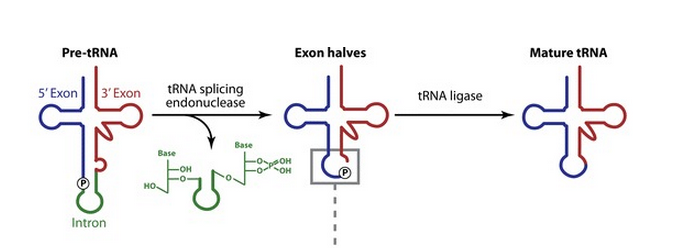
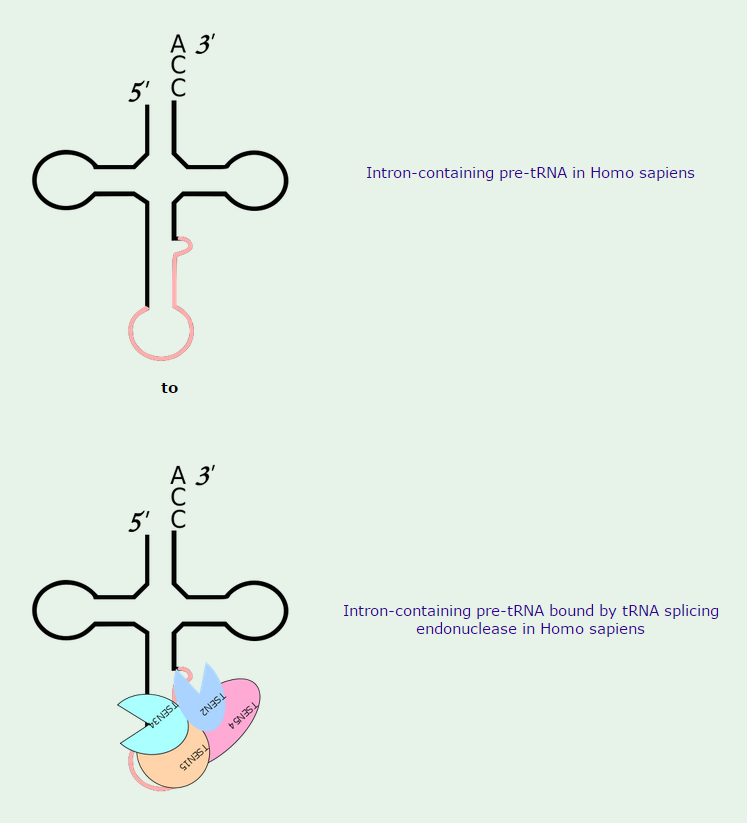
Like most other eucaryotic RNAs, tRNAs are covalently modified before they are allowed to exit from the nucleus. Eucaryotic tRNAs are synthesized by RNA Polymerase III. Both bacterial and eucaryotic tRNAs are typically synthesized as larger precursor tRNAs, which are then trimmed to produce the mature tRNA. In addition, some tRNA precursors (from both bacteria and eucaryotes) contain introns that must be spliced out. This splicing reaction differs chemically from pre-mRNA splicing; rather than generating a lariat intermediate, tRNA splicing uses a cut-and-paste mechanism that is catalyzed by proteins See below:
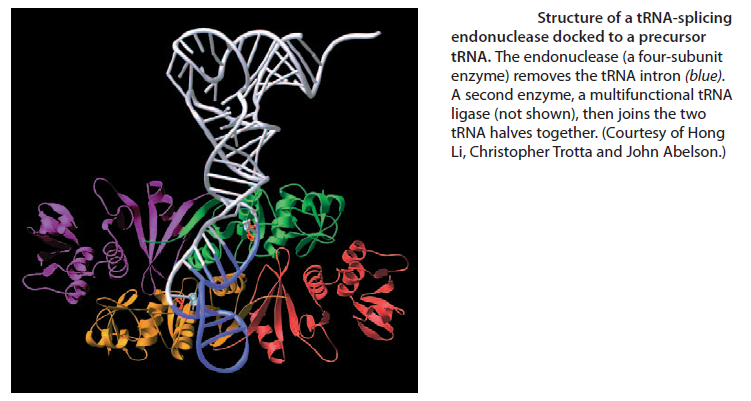
Trimming and splicing both require the precursor tRNA to be correctly folded in its cloverleaf configuration. Because misfolded tRNA precursors will not be processed properly, the trimming and splicing reactions are thought to act as quality- control steps in the generation of tRNAs. All tRNAs are modified chemically—nearly 1 in 10 nucleotides in each mature tRNA molecule is an altered version of a standard G, U, C, or A ribonucleotide.Over 50 different types of tRNA modifications are known; a few are shown below:
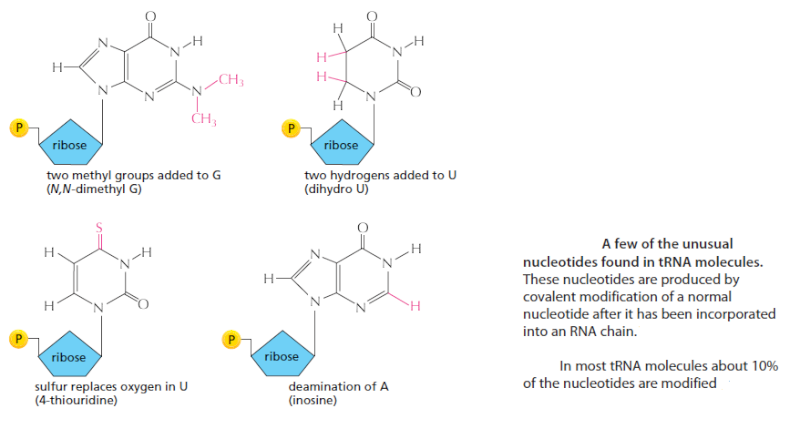
Some of the modified nucleotides—most notably inosine, produced by the deamination of adenosine—affect the conformation and basepairing of the anticodon and thereby facilitate the recognition of the appropriate mRNA codon by the tRNA molecule. Others affect the accuracy with which the tRNA is attached to the correct amino acid.Some of the modified nucleotides—most notably inosine, produced by the deamination of adenosine—affect the conformation and basepairing of the anticodon and thereby facilitate the recognition of the appropriate mRNA codon by the tRNA molecule . Others affect the accuracy with which the tRNA is attached to the correct amino acid.[/b]
After tRNA is transcribed by RNA polymerase III as a precursor tRNA, it must be processed into a mature tRNA
This happens through the removal, addition and chemical modification of nucleotides. Processing for some tRNA involves 2
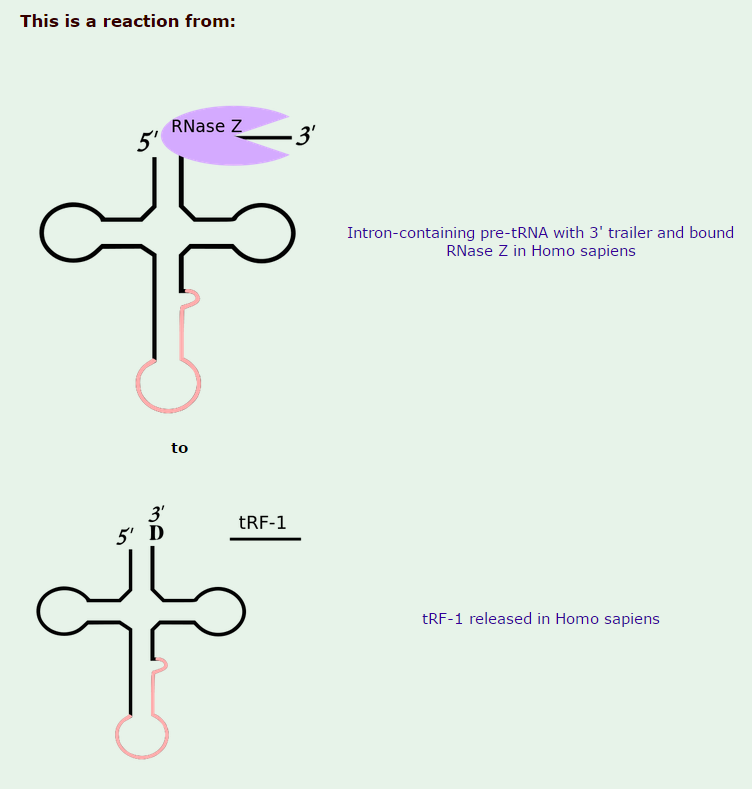
1) removal of the leader sequence at the 5 prime end
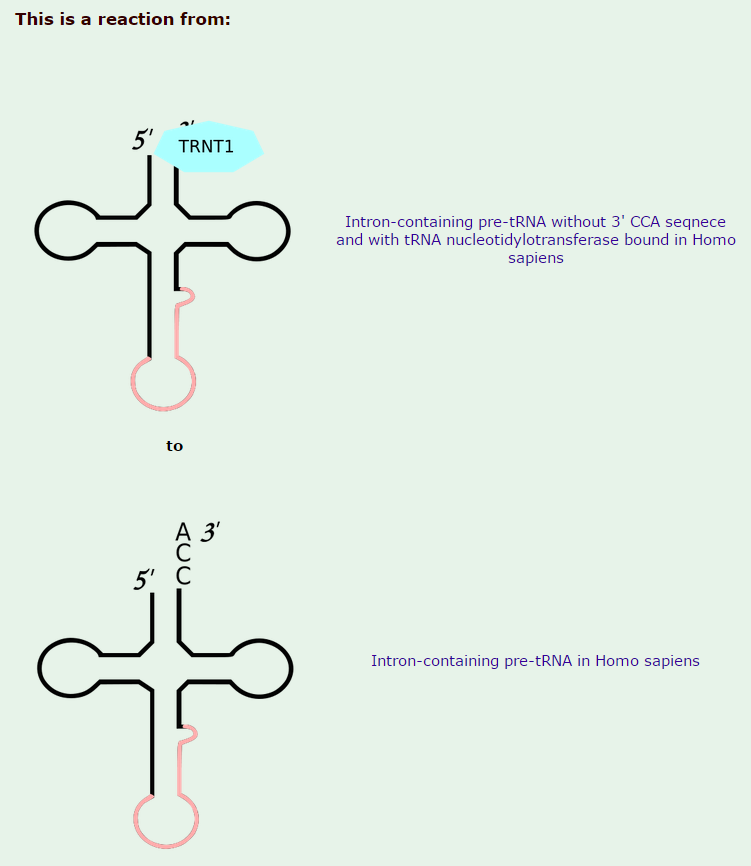
2) replacement of two nucleotides at the 3 prime end by the sequence CCA (with which all mature tRNA molecules terminate)
3) chemical modification of certain bases and
4) excision of an intron.
The mature tRNA is often diagrammed as a flattened cloverleaf which clearly shows the base pairing between self-complementary stretches in the molecule.
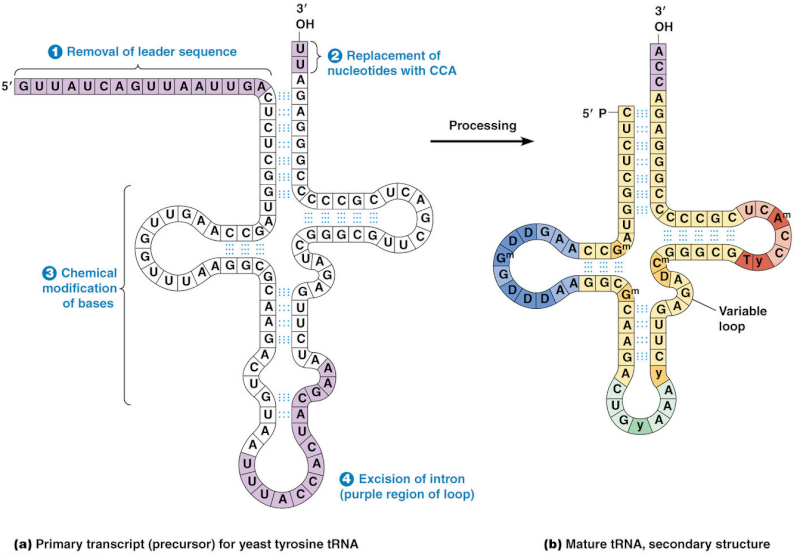
tRNA maturation in Homo sapiens
Enzymatic complexes involved in the process: 18
Proteins:
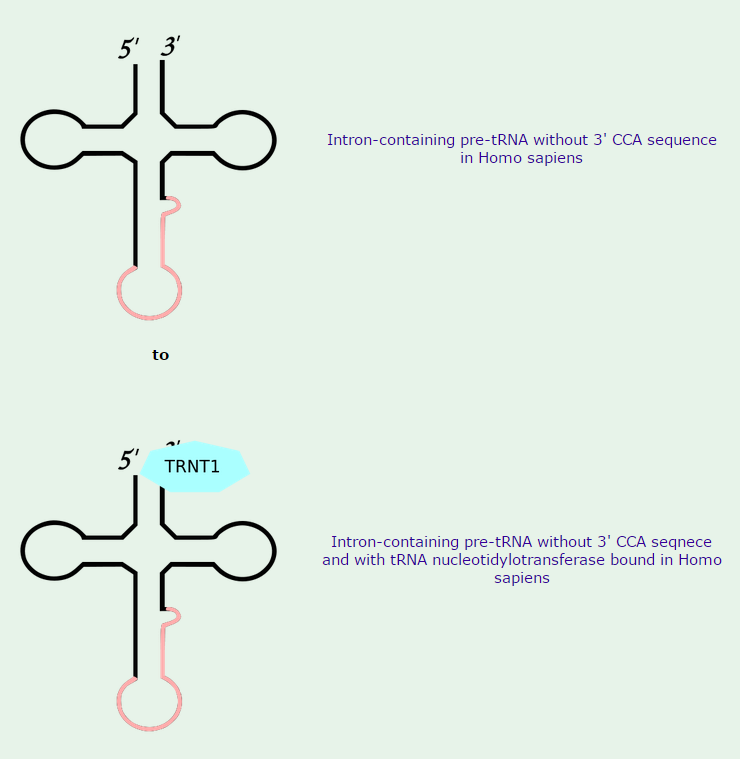
CCA tRNA nucleotidyltransferase 1
Zinc phosphodiesterase ELAC protein 2
Enzymatic complexes:
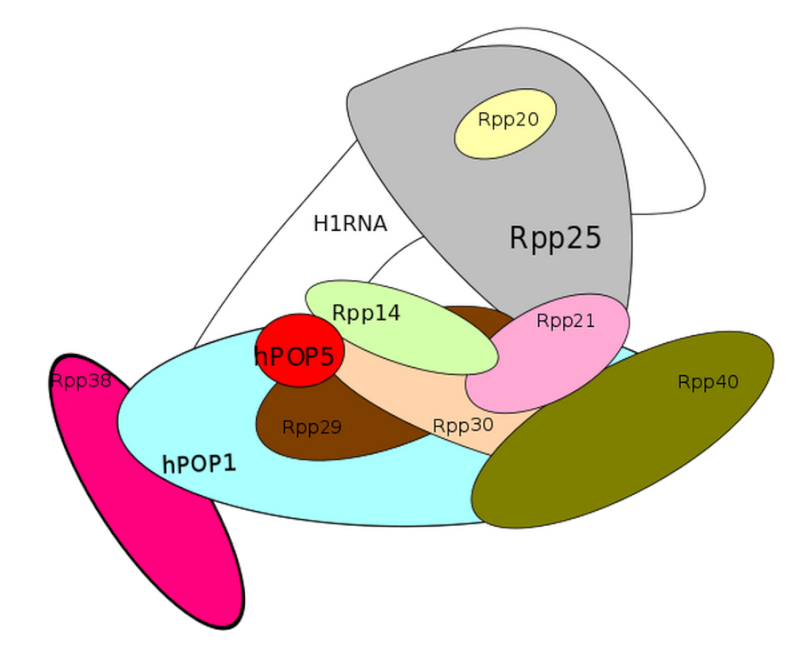
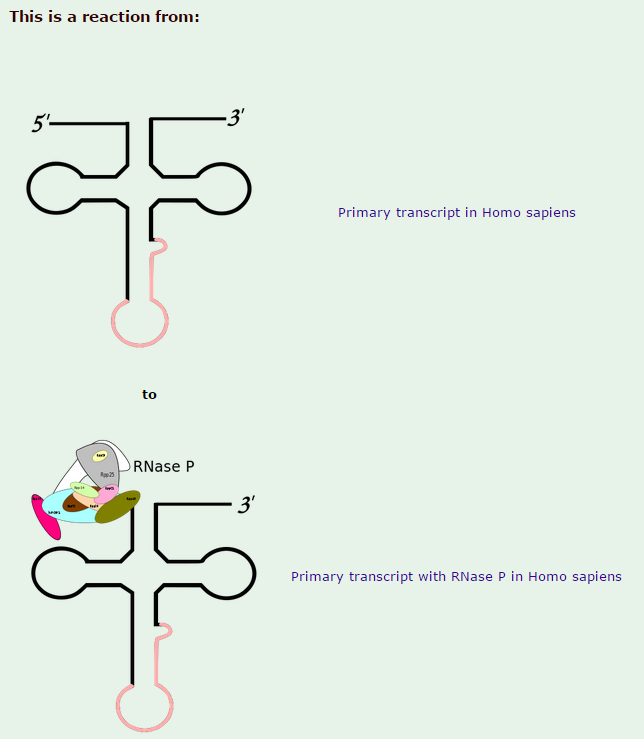
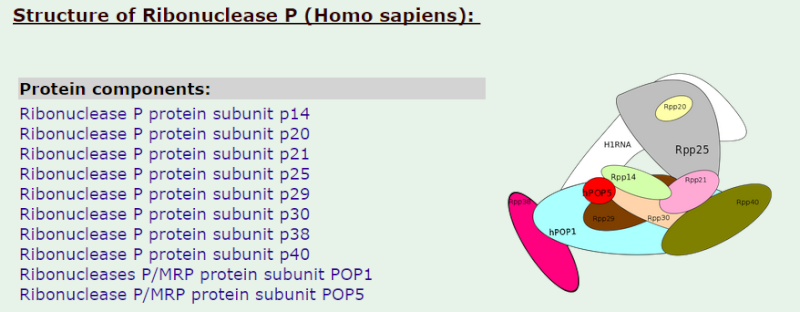
Ribonuclease P
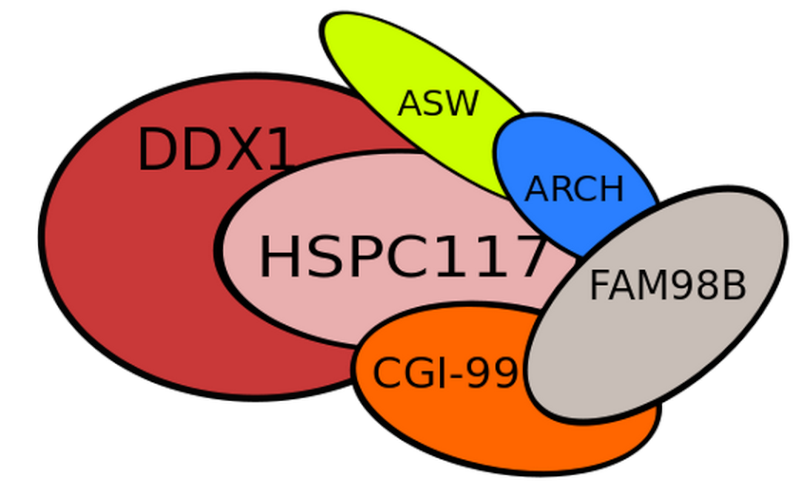
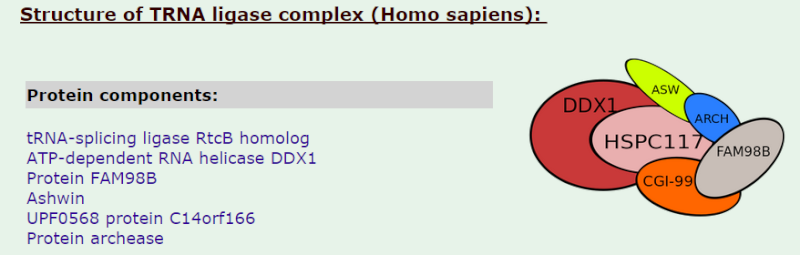
tRNA ligase complex
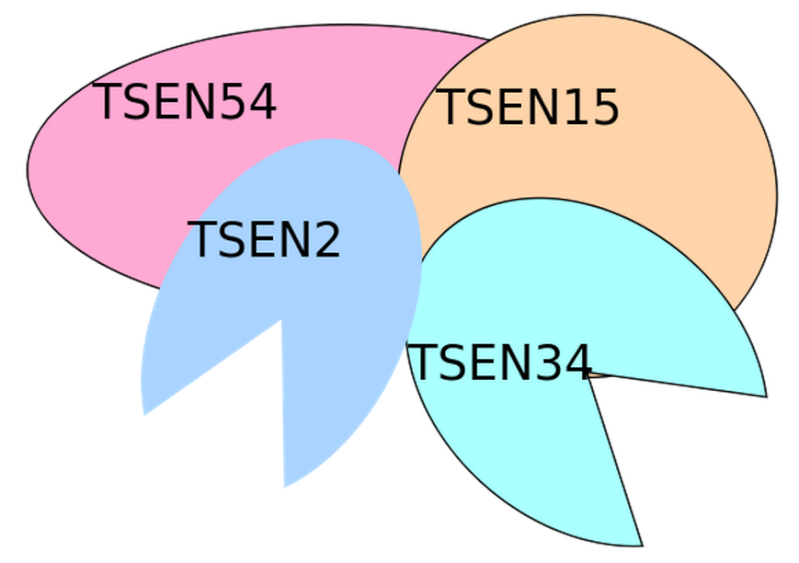
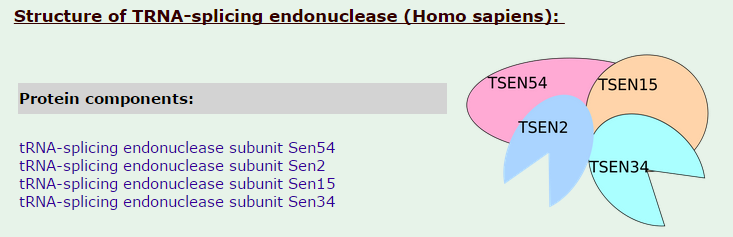
tRNA-splicing endonuclease
17
19
 16
16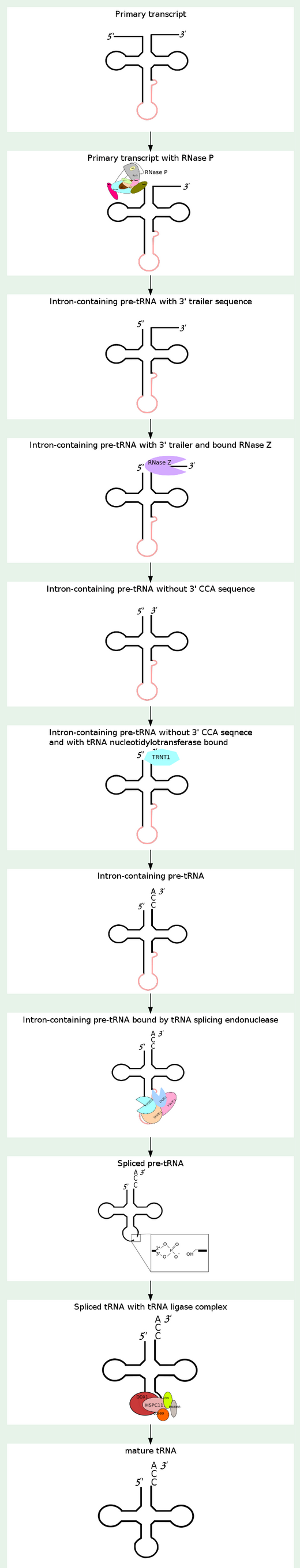
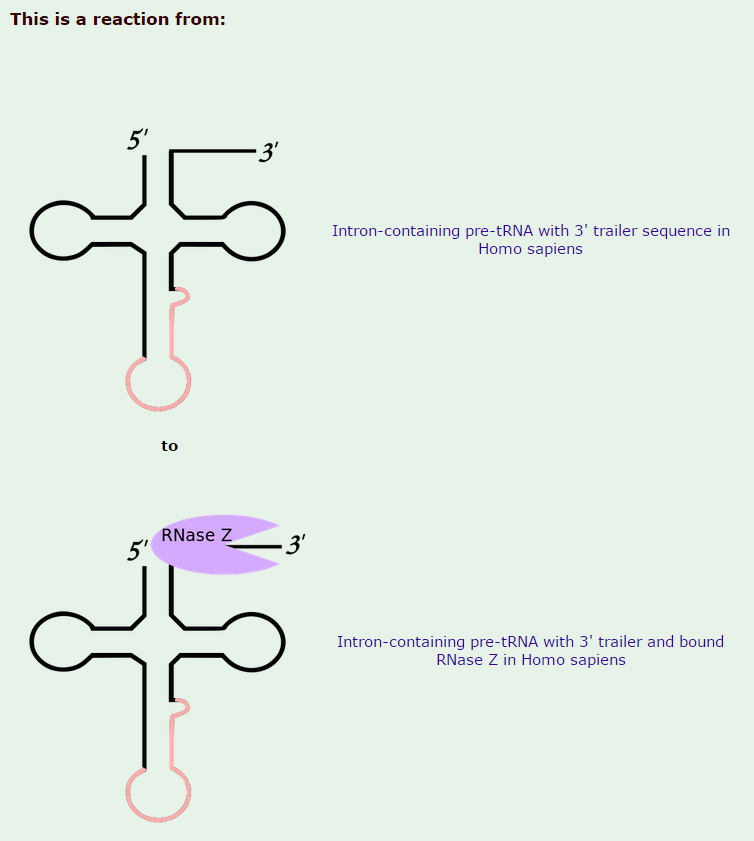
1)The transcription product, the pre-tRNA, contains additional RNA sequences at both the 5’ and 3’-ends. These additional sequences are removed from the transcript during processing. The additional nucleotides at the 5’-end are removed by an unusual RNA containing enzyme called ribonuclease P (RNase P)
2.
Some tRNA precursors contain an intron located in the anticodon arm. These introns are spliced out during processing of the tRNA.
The cloverleaf structure of a single polynucleotide tRNA molecule is universally conserved among organisms. However, tRNA genes are often divided into parts on the chromosome; in bacteria, archaea, eukarya, and organelles, several tRNA genes are interrupted by various types of introns, which are removed by RNA splicing after transcription
Introns in nuclear and archaeal tRNAs are generally cleaved by tRNA-splicing endonuclease 13
tRNA splicing is a fundamental process required for cell growth and division. SEN2 is a subunit of the tRNA splicing endonuclease, which catalyzes the removal of introns, the first step in tRNA splicing 14
The tRNA splicing reaction in yeast occurs in three steps; each step is catalyzed by a distinct enzyme, which can function interchangeably on all of the substrates 15
3.
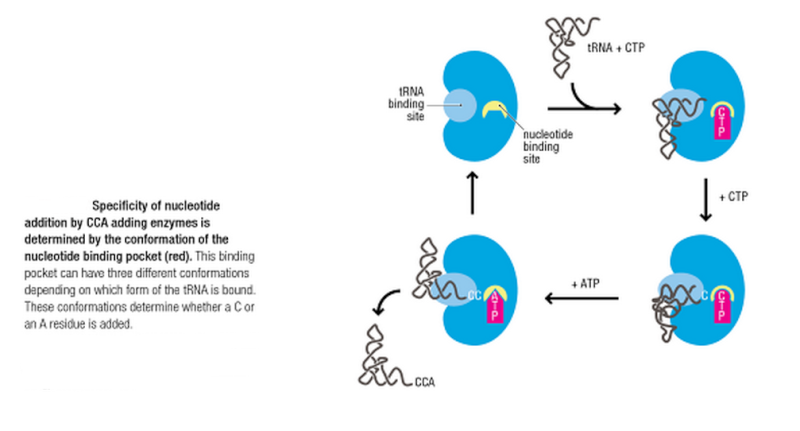
All mature tRNAs contain the trinucleotide CCA at their 3’-end. These three bases are not coded for by the tRNA gene. Instead, these nucleotides are added during processing of the pre-tRNA transcript. The enzyme responsible for the addition of the CCA-end is tRNA nucleotidyl transferase and the reaction proceeds according to the following scheme:
tRNA +CTP --> tRNA-C + PPi (pyrophosphate)
tRNA-C +CTP --> tRNA-C-C + PPi
tRNA-C-C +ATP --> tRNA-C-C-A + PPi
4.
Mature tRNAs can contain up to 10% bases other than the usual adenine (A), guanine (G), cytidine (C) and uracil (U). These base modifications are introduced into the tRNA at the final processing step. The biological function of most of the modified bases is uncertain and the translation process seems normal in mutants lacking the enzymes responsible for modifying the bases.
Termination signals end the transcription of RNA by RNA polymerase I and RNA polymerase III without the activity of hairpin structures as seen in prokaryotes.
mRNA is cleaved 10 to 35 base-pairs downstream of a AAUAAA sequence (which acts as a poly-A tail addition signal).
The biogenesis of mature transfer (t)RNAs in cells has a complexity that belies the elegance of their function as the adaptor molecules of protein synthesis. They are synthesized as precursors which are converted to mature tRNA molecules by a sequence of events that includes processing of their 5′ and 3′ ends, modification of a number of bases, addition of the terminal CCA residues and, in the case of intron-containing tRNAs, splicing 1
Transfer RNAs (tRNAs) play an important role linking mRNA and amino acids during protein biogenesis. Four types of tRNA genes have been identified in living organisms. However, the evolutionary origin of tRNAs remains largely unknown. Given their central role in life and their high level of conservation, tRNAs have stimulated extensive interest in their origin and evolution
This enzyme alone is like a small cogwheel in a watch in a complex clockwork mechanism. If you take it away, the whole mechanism of protein synthesis ceases to exist. No RNase, no protein synthesis, no life....... As with RNase, the Protein production needs hundreds, well, probably thousands of intricate fine-tuned parts, all doing their function in a precise way, and if you take away just one small apparently irrelevant part, bye-bye proteins, bye-bye life. These enzyme subparts have no use by their own, but only if correctly inserted in the intricate holoenzyme complexes exercising precisely their function.........
further readings :
http://journal.frontiersin.org/researchtopic/1729/molecular-biology-of-the-transfer-rna-revisited
Molecular biology of the transfer RNA revisited
Last edited by Otangelo on Sun Sep 04, 2022 2:13 pm; edited 66 times in total



 ´
´