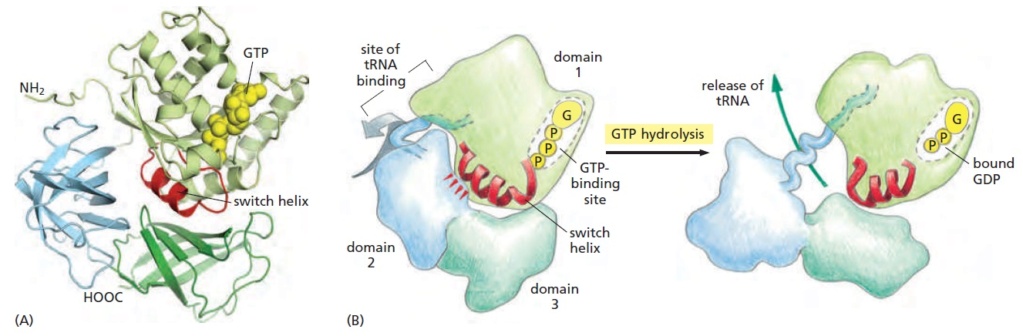
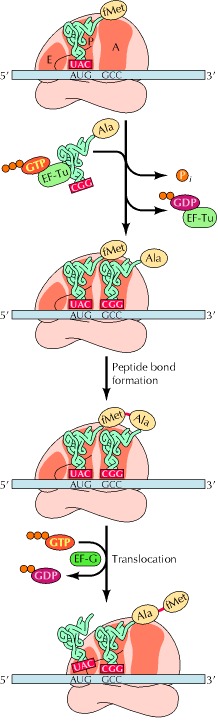
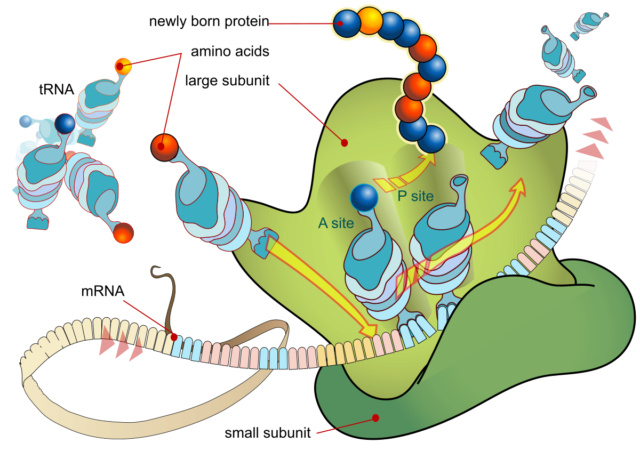
The ribosome is composed of a large and a small subunit, which are assembled on the translation initiation region (TIR) of the mRNA during the initiation phase of translation.
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC1082788/
My comment "Know-how", foreknowledge, and goal orientedness is necessary for this machine-like process to occur. It has to be pre-programmed. Unguided, random events would never "know" that the small and large ribosomal subunits have to be assembled at the translation initiation region (TIR) for the initiation phase of translation to occur. There is simply no requirement for natural selection or any proposed mindless process to instantiate, program, and orchestrate such a complex precise process.
The problem is that nature has too many options and without design couldn’t/wouldn't sort them all out. Natural mechanisms are too unspecific, chaotic, random, and without goal to determine any particular outcome. Natural processes could hypothetically form a machine-like assembly process, but also compatible with the formation of a wide variety of other molecular assemblages, most of which have no biological relevance or function. Random molecules have a total freedom of arrangements and chemical reactions. Yet it’s precisely that randomness that makes chemicals unable to account for specific mechanistic arrangements of remote possibility to happen. Chemicals without intent rather than resulting in structured functional ordered assembly of complex mechanistic structres give any non-functional outcome, and lead to doing anything. Yet when one of those things is a highly improbable specified event, design is the more case-adequate explanation. Occam's razor also boils down to an argument from ignorance: in the absence of better information, you use a heuristic to accept one hypothesis over the other.
In the following elongation phase, the mRNA is decoded as it slides through the ribosome and a polypeptide chain is synthesized. Elongation continues until the ribosome encounters a stop codon on the mRNA and the process enters the termination phase of protein synthesis. Newly synthesized protein is released from the ribosome. In the final ribosome recycling phase, the ribosomal subunits dissociate and the mRNA is released. Each phase is regulated by a number of different factors. Reviews of the phases are available.
My comment: Keeping a stable internal cellular environment requires constant adjustments as conditions change inside and outside the cell. The adjusting of systems within a cell is called homeostatic regulation. Because the internal and external environments of a cell are constantly changing, adjustments must be made continuously to stay at or near the set point (the normal level or range).
Regulation is ubiquitous in biology. Regulation of biological processes occurs when any process is modulated in its frequency, rate or extent. Biological processes are regulated by many means; examples include the control of gene expression, protein modification or interaction with a protein or substrate molecule. While Gene expression in prokaryotes is regulated primarily at the transcriptional level, in eukaryotic cells, it is regulated at many levels (epigenetic, transcriptional, post-transcriptional, translational, and post-translational). Metabolism or intracellular signaling also requires regulation. Gene regulation is a dynamical process composed of a number of steps, for example the binding of Transcription Factors to DNA, recruitment of transcription machinery and the production of the messenger RNA, post-transcriptional regulation, splicing and transport of mRNA, translation, maturation and possible localization of proteins. Alternative splicing is now understood to be a common mechanism of gene regulation in eukaryotes; according to one estimate, 70% of genes in humans are expressed as multiple proteins through alternative splicing. The primary method to control what type and how much protein is expressed in a prokaryotic cell is through the regulation of DNA transcription into RNA.
Biological regulation: controlling the system from within 06 August 2015
Regulation requires a distinctive architecture of functional relationships, and specifically the action of a dedicated subsystem whose activity is dynamically decoupled from that of the constitutive regime.
https://link.springer.com/article/10.1007/s10539-015-9497-8
Properties of Life
All living organisms share several key characteristics or functions: order, sensitivity or response to the environment, reproduction, growth and development, regulation, homeostasis, and energy processing. When viewed together, these characteristics serve to define life.
https://courses.lumenlearning.com/wm-biology2/chapter/properties-of-life/
Regulate means control or maintain the rate or speed of (a machine or process) so that it operates properly. Regulation is the management of complex systems according to a set of rules and trends. It is a rule or directive made and maintained by an authority.
Regulating is always either a) a preprogrammed action by an intelligent agency, or b) an action where an intelligent agent is directly actively involved. Setting rules, giving directives for specific purposes or functions is always the result of a mental action. It is logical to conclude that biological regulation requires intelligent set up.
Regulation is essential in biology
1. Regulation is ubiquitous in biology, and a fundamental property of living systems. Keeping a stable internal cellular environment requires constant adjustments as conditions change inside and outside the cell. The adjusting of systems within a cell is called homeostatic regulation. Because the internal and external environments of a cell are constantly changing, adjustments must be made continuously to stay at or near the set point (the normal level or range). Regulation of biological processes occurs when any process is modulated in its frequency, rate or extent. Biological processes are regulated by many means; examples include the control of gene expression, protein modification or interaction with a protein or substrate molecule. While Gene expression in prokaryotes is regulated primarily at the transcriptional level, in eukaryotic cells, it is regulated at many levels (epigenetic, transcriptional, post-transcriptional, translational, and post-translational). Metabolism or intracellular signaling also requires regulation. Gene regulation is a dynamical process composed of a number of steps, for example the binding of Transcription Factors to DNA, recruitment of transcription machinery and the production of the messenger RNA, post-transcriptional regulation, splicing and transport of mRNA, translation, maturation and possible localization of proteins.
2. Regulation requires a distinctive architecture of functional relationships, and specifically the action of a dedicated subsystem whose activity is dynamically decoupled from that of the constitutive regime. Regulate means control or maintain the rate or speed of (a machine or process) so that it operates properly. Regulation is the management of complex systems according to a set of rules and trends. It is a rule or directive made and maintained by an authority. Regulating is always either a) a preprogrammed action by an intelligent agency, or b) an action where an intelligent agent is directly actively involved.
3. Setting rules, giving directives for specific purposes or functions is always the result of a mental action. It is logical to conclude that biological regulation requires intelligent set up.
Although the main events of the translation process are universally conserved, major differences in the detailed mechanism of each phase exist. Bacterial translation involves relatively few factors, in contrast to the more complex process in eukaryotes . Here we focus on translation initiation in bacteria. Archaeal and eukaryotic processes of translation initiation are reviewed elsewhere.
BACTERIAL TRANSLATION INITIATION
Ribosomes initiate translation on mRNAs already during transcription. Hence, transcription and translation are tightly coupled cellular processes. Translation initiation is the rate-limiting and most highly regulated phase of the four phases in protein biosynthesis.
The rate at which ribosomes assemble on the mRNA is on the order of seconds, although it is specific for each mRNA. The ribosomes subsequently translate the mRNA at a rate of approximately 12 amino acids per second . The ribosome, the aminoacylated and formylated initiator tRNA (fMet-tRNAfMet), mRNA, and the three protein factors, initiation factor IF1, initiation factor IF2, and initiation factor IF3, are involved in the translation initiation phase

Translation initiation pathway in bacteria.
The 30S and 50S ribosomal subunits are shown in light and dark grey, respectively. Translation initiation factors IF1, IF2, and IF3, the mRNA, and the fMet-tRNAfMet are shown in red, blue, green, yellow, and magenta, respectively. The components are placed on the ribosome according to current experimental knowledge. Details of the pathway are given in the text. Structures are derived from PDB entries as follows: 30S ribosomal subunit, 1HR0; 50S ribosomal subunit, 1FFK; IF1, 1HR0; IF2, 1G7T; IF3N, 1TIF; IF3C, 1TIG; mRNA, 1JGQ; fMet-tRNAfMet, 1JGQ.
The bacterial 70S ribosome is composed of a large 50S and a small 30S subunit. It has three tRNA binding sites designated the aminoacyl (A), peptidyl (P), and exit (E) sites. Binding of IF3 to the 30S ribosomal subunit promotes dissociation of the ribosome into subunits and thus couples ribosome recycling and translation initiation. Initiation factor IF1 binds specifically to the base of the A-site of the 30S ribosomal subunit and is thought to direct the initiator tRNA to the ribosomal P-site by blocking the A-site. IF1 stimulates the activities of IF3 and hence also the dissociation of the ribosomal subunits.
Following subunit dissociation, IF2, mRNA, and fMet-tRNAfMet associate with the 30S ribosomal subunit in an unknown and possibly random order. The Shine-Dalgamo (SD) sequence of canonical mRNAs interacts with the anti-SD sequence of the 16S rRNA, and the initiation codon is adjusted in the P-site of the ribosome. The initiation factors (especially IF3) seem to be responsible for this adjustment. The initiator tRNA is positioned in the P-site of the 30S ribosomal subunit in three steps that are designated codon-independent binding, codon-dependent binding, and fMet-tRNAfMet adjustment. All three steps are probably promoted by IF2, which interacts with fMet-tRNAfMet on the ribosome. Furthermore, IF3 stabilizes the binding of fMet-tRNAfMet to the ribosomal P-site and confers proofreading capability by destabilization of a mismatched codon-anticodon interaction.
The 30S preinitiation complex consists of the 30S ribosomal subunit, the three initiation factors, and mRNA in a standby position where fMet-tRNAfMet is bound in a codon-independent manner. This relatively unstable complex undergoes a rate-limiting conformational change that promotes the codon-anticodon interaction and forms the more stable 30S initiation complex. Initiation factors IF1 and IF3 are ejected, while IF2 stimulates association of the 50S ribosomal subunit to the complex. Initiator fMet-tRNAfMet is adjusted to the correct position in the P-site, and IF2 is released from the complex. During this process, GTP bound to IF2 is hydrolyzed to GDP and Pi. The newly formed 70S initiation complex holding fMet-tRNAfMet as a substrate for the peptidyltransferase center of the 50S ribosomal subunit is ready to enter the elongation phase of translation.
COMPONENTS INVOLVED IN TRANSLATION INITIATION
The translation initiation event is a complex and highly regulated process involving both RNA and protein components.
Stabilization of the ribosomal structure.
Two-thirds of the ribosome is composed of RNA. Three main types of interactions stabilize the tertiary structure of the rRNA:
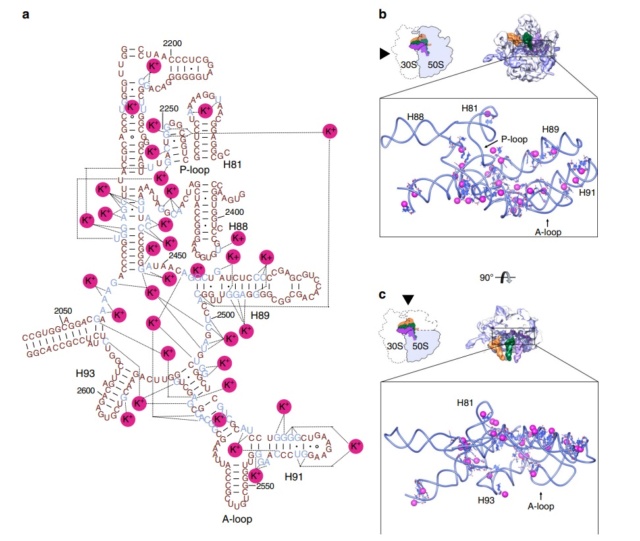
(i) Mg2+ bridges,
(ii) RNA-RNA interactions, and
(iii) RNA-protein interactions.
The magnesium ions form neutralizing bridges between two or more phosphate groups from secondary-structure elements remote in sequence. RNA-RNA interactions of different types exist: (i) base pairing between nucleotides associated with secondary-structure elements remote in sequence, and (ii) A-minor motifs. The A-minor motif is an interaction between an adenine that inserts its minor groove face into the minor groove of a base pair in a helix. This is most often a GC pair. The adenine forms hydrogen bonds with one or both of the backbone 2′ hydroxyl groups of the RNA duplex. Different types of helix-helix packing interactions occur, involving the insertion of a ridge of phosphates into the minor groove of another helix or using an unpaired purine base to mediate the perpendicular packing of one helix against the minor groove of another.
RNA-protein interactions occur mainly via the sugar-phosphate backbone of the RNA. Thus, the ribosomal proteins recognize the unique shape of the rRNA rather than the bases, and the interactions are therefore sequence unspecific. Many of the ribosomal proteins have nonglobular extensions that are highly conserved in sequence. These tails penetrate into the ribosome and fill the gaps between RNA helices. In isolation, these protein tails, which contain approximately 26% arginine and lysine residues, look like random coils that probably only assume the conformation they have on the ribosome when bound (4, 219).
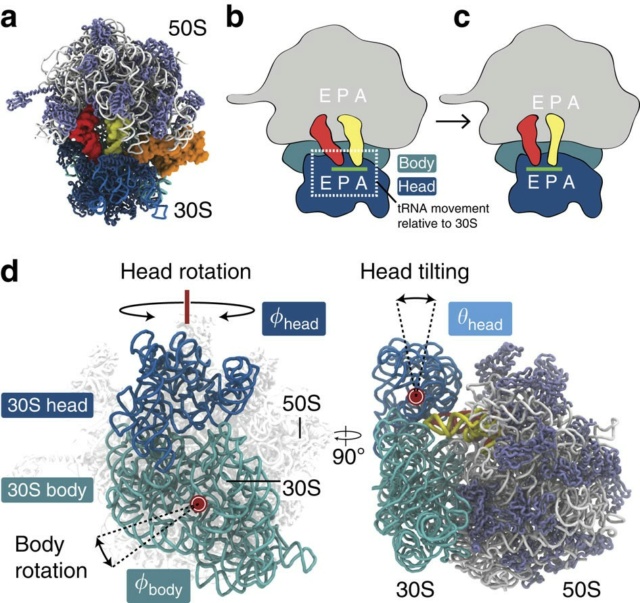
Prior to peptide bond formation, an aminoacyl-tRNA is bound in the ribosomal A-site, a peptidyl-tRNA is bound in the P-site, and a deacylated tRNA, which is ready for ejection from the ribosome, is bound to the E-site. Translation moves the tRNA from the A-site through the P- and E-sites before they exit the ribosome again, with the exception of the initiator tRNA, which binds directly to the P-site. The small ribosomal subunit contains the decoding center, where the triplet codons of the mRNA are base-paired with the anticodons of the cognate tRNA, and hence determines the sequence of amino acids to be incorporated in the synthesized protein. The large subunit contains the peptidyltransferase center and is thus the catalytic unit.
Small ribosomal subunit
The small ribosomal subunit is composed of 21 proteins and an RNA of approximately 1,500 nucleotides sedimenting at 16S. The shape of the subunit is determined largely by the RNA component, which forms four secondary-structure domains (Fig. (Fig.2)2).

Structures of the ribosomal subunits.
(A) Overview of the 16S rRNA secondary structure. The domains are shown in colors according to the secondary structure: blue, 5′ domain (bulk of body); magenta, central domain (platform); red, 3′ major domain (head); yellow, 3′ minor domain (helices 44 and 45 located at the subunit interface).
(B) Overview of the 23S and 5S rRNA secondary structures. The RNA domains are shown in colors according to the secondary structure of the 23S rRNA: blue, domain I; cyan, domain II; green, domain III; yellow, domain IV; red, domain V; magenta, domain VI. The 5S rRNA is shown in orange.
(C) Three-dimensional structure of the 30S ribosomal subunit from T. thermophilus at 3-Å resolution (PDB entry 1J5E). RNA secondary-structure domains are colored as in panel A. Note that the secondary-structure domains of the RNA correspond well to the tertiary domains. Proteins are omitted for clarity. The tRNA binding sites A, P, and E are indicated.
(D) Three-dimensional structure of the 50S ribosomal subunit from H. marismortui at 2.4-Å resolution (PDB entry 1FFK). Colors are the same as in panel B. Note that the secondary-structure domains of the RNA do not correspond to the tertiary domains, unlike for the 30S subunit. Proteins are omitted for clarity. The L1 stalk, the central protuberance (CP), and the L7-L12 stalk are indicated.
Traditionally, the subunit has been divided into an upper third, called the head, connected by the neck to the body with a shoulder and platform. A protrusion in the lower part of the body is called the spur (or toe). The side of the 30S subunit facing the 50S subunit is called the front, whereas the solvent-exposed side is called the back. The small subunit binds mRNA and the anticodon loop and stem of tRNAs. Translational fidelity is controlled on the subunit by monitoring the base pairing between the codon and anticodon in the process known as decoding. The decoding center located at the upper part of the body and lower part of the head of the subunit is constructed entirely of RNA and contains, among other elements, the upper part of helix 44 and the 3′ and 5′ ends of the 16S rRNA. An interaction that is important for translation initiation occurs at the 3′ end of the 16S rRNA (also known as the anti-SD [ASD] sequence) that base-pairs with the SD sequence of the mRNA.
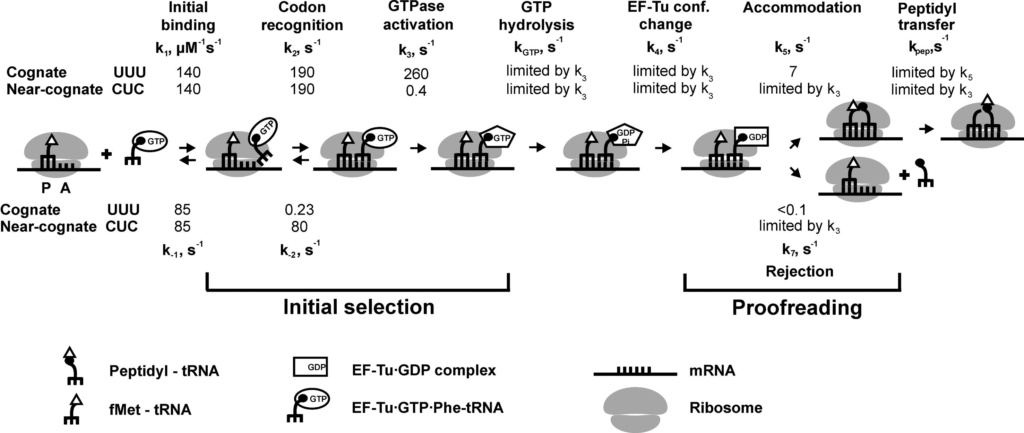
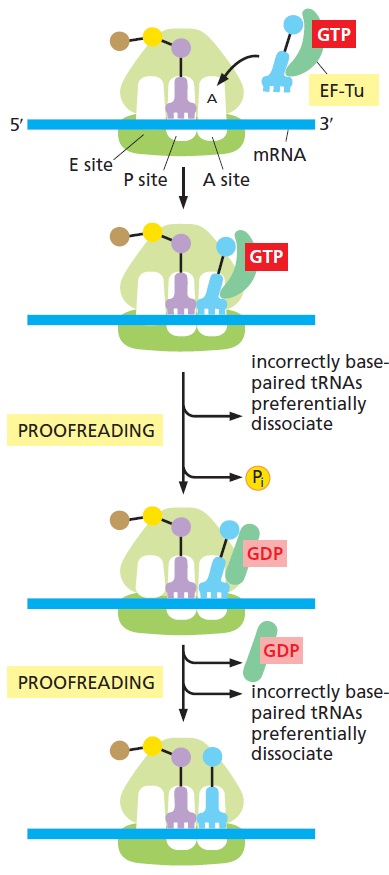
E. coli has 41 different tRNA species with different anticodons. The ribosome must select the tRNA with an anticodon complementary to the codon of the mRNA. This is termed the cognate tRNA. The error rate of tRNA selection in the decoding process is 1000 to 10.000. In addition to having lower dissociation rates from the ribosome, cognate tRNAs have higher rates of elongation factor EF1A GTPase activation and accommodation (movement of the aminoacyl end of tRNA into the A-site of the 50S ribosomal subunit) than do near-cognate tRNAs. Based on this result, it was proposed that binding of cognate tRNA induces a conformational change of the ribosome. Crystal structures of the 30S subunit complexed with mRNA and cognate tRNA in the A-site reveal an induced fit mechanism. Bases A1492 and A1493 of the 16S rRNA flip out of helix 44 and interact with the correctly base-paired codon-anticodon helix in an A-minor motif type interaction. A1492 and A1493 interact with the minor groove of a correctly paired codon-anticodon but not with incorrectly paired codons and anticodons. Binding of cognate tRNA also causes a flip of G530 of the 16S rRNA from syn to anti conformation. Bases A1492 and A1493 interact with the first and second base pairs of the codon-anticodon helix, respectively. G530 interacts with the second position of the anticodon as well as the third position of the codon. The result is a strictly monitored codon-anticodon interaction in the first two positions, whereas the ribosome is able to tolerate noncanonical base pairs at the third position. During decoding, the flipping of the 30S subunit bases A1492, A1493, and G530 translates to other parts of the subunit and leaves it in a closed conformation in which the shoulder and head domains are rotated toward the subunit center, compared to the more open structure when the A-site is unoccupied. The transition to the more closed state is unfavorable for near-cognate tRNAs. However, X-ray crystal structures of the intact 70S E. coli ribosome reveal that the closing of the head domain of the 30S subunit is connected to formation of the intact 70S ribosome and not to decoding. These structures do, however, indicate a movement of the small-subunit body connected with decoding. Thus, ribosomes play an active role in tRNA selection by direct recognition of the codon-anticodon base pairing.
Large ribosomal subunit.
The large ribosomal subunit is composed of 34 proteins and two RNAs sedimenting at 5S and 23S, containing about 120 and 2,900 nucleotides, respectively. Six secondary-structure domains are defined by the 23S rRNA, whereas the 5S rRNA is regarded as the seventh domain of the subunit. A direct relationship between secondary structure elements and morphological domains is not present in the large subunit, which presents a more compact structure than the small subunit (Fig. (Fig.2).2). The 50S subunit consists of a rounded base with three protuberances called the L1 protuberance, the central protuberance, and the L7/L12 stalk (Fig. (Fig.2)2). A tunnel starts at the peptidyltransferase center (PTC), where the formation of peptide bonds occurs. The nascent polypeptide is thought to exit at the base of the cytoplasmic side of the subunit through the approximately 100-Å-long tunnel, which has an average diameter of 15 Å.
During the peptidyl transfer reaction, the α-amino group of A-site tRNA attacks the carbonyl group of the P-site peptidyl group, which is linked to the tRNA via an ester bond. The reaction proceeds via a tetrahedral intermediate to form a peptidyl bond. The reaction occurs in the PTC, where the amino acid of the A-site tRNA has been properly positioned relative to the nascent peptide chain bound to the P-site tRNA. Peptide bond formation is then catalyzed. The PTC was identified by soaking crystals of Haloarcula marismortui 50S subunits with a transition state analogue, the so-called Yarus inhibitor. Surprisingly, the subunit is completely devoid of protein within 18 Å of the PTC, and the ribosome is thus a ribozyme. It is beyond the scope of this review to go into detail about the mechanism of the peptidyl transferase reaction.
After the peptidyltransferase reaction has occurred, a deacylated tRNA is left in the P-site and the A-site tRNA is covalently bound to a peptide chain extended by one residue. For elongation to proceed, the P-site tRNA has to move into the E-site ready for ejection from the ribosome and the A-site peptidyl-tRNA has to move to the P-site. The E-site is specific for deacylated tRNAs. The movement of tRNAs must be accompanied by a precise movement of the mRNA to preserve the reading frame.
mRNA
mRNA interacts specifically with tRNA as well as the 30S ribosomal subunit during translation initiation. The mRNA covered by the ribosome in the translation initiation phase is called the ribosomal binding site (RBS) and extends over about 30 nucleotides. Bacterial mRNAs are normally polycistronic and possess multiple signals for initiation and termination of protein synthesis.
TIRs on mRNAs are not only characterized by the presence of a putative initiation codon. Additional elements are necessary to promote correct initiation and avoid initiation from, for instance, AUG codons encoding internal methionines of a protein. Upstream from the initiation codon is the 5′ untranslated region (5′ UTR). This region contains the SD sequence, which can undergo base-pairing to the 3′ of the 16S rRNA of the 30S ribosomal subunit. A direct consequence of the SD interaction is the adjustment of the initiation codon to the ribosomal P-site, where it interacts with fMet-tRNAfMet. E. coli mRNAs typically have the SD sequence GGAGG located 7 ± 2 nucleotides upstream from the initiation codon, which can be AUG, GUG or UUG. The exceptional AUU initiation codon has been observed in infC (encoding IF3) and pcnB [encoding E. coli poly(A) polymerase]. Initiation codons in E. coli occur at a frequency of 90, 8, and 1% for AUG, GUG, and UUG, respectively.
Ribosomal protein S1 interacts with a pyrimidine-rich region 5′ to the SD region on mRNAs. This pyrimidine-rich region acts as a ribosome recognition site. A direct interaction has been confirmed by cryoelectron microscopy (EM) studies of S1 on the 30S ribosomal subunit with a bound mRNA.
A region downstream from the initiation codon of several mRNAs was found to show complementarity to bases 1469 to 1483 within helix 44 of the 16S rRNA. This region was named the downstream box (DB), and there appeared to be a correlation between the degree of complementarity to the 16S rRNA and the translational efficiency of the mRNA. A mechanism similar to the SD-ASD interaction was proposed. The presence of a DB-anti-DB (ADB) interaction has been a matter of debate, and a recent review concludes that there is no biochemical or genetic evidence in support of the proposed role of the DB-ADB interaction in ribosomal recruitment of mRNA. This is supported by the crystal structure of the T. thermophilus 30S ribosomal subunit, which reveals that the shoulder of the subunit is located between the putative DB of the mRNA and the proposed anti-DB of the 16S rRNA .
Bacterial mRNAs are either canonical or leaderless, although the latter is rare, with no more than ∼40 identified cases in bacteria. Canonical mRNAs contain the 5′ UTR elements described above, whereas leaderless mRNAs start at, or a few nucleotides 5′ upstream of, the initiation codon. A clear mechanism for binding of canonical mRNAs and the order in which mRNA and fMet-tRNAfMet enter the 30S ribosomal subunit has not been established (Fig. (Fig.3).3).
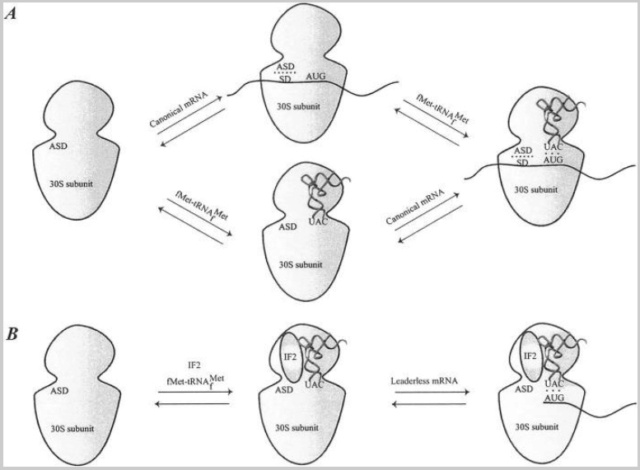
Binding of mRNA to the 30S ribosomal subunit.
(A) Binding of a canonical mRNA to the 30S ribosomal subunit. Two alternative pathways are shown where either the mRNA or the fMet-tRNAfMet binds first, followed by the other component. The mRNA is bound via the SD-ASD interaction as well as the codon-anticodon interaction.
(B) Binding of a leaderless mRNA to the 30S ribosomal subunit. The mRNA is bound to the ribosome mainly via the codon-anticodon interaction. IF2 stimulates the binding of leaderless mRNAs, presumably by recruitment of fMet-tRNAfMet to the subunit.
Initiation factors do not affect the SD-ASD interaction or the association between the 30S ribosomal subunit and canonical mRNA (61). However, site-directed cross-linking experiments have shown that mRNA is partially relocated on the 30S ribosomal subunit from a “standby site” to a site closer to the P-site in a process influenced by IF1, IF2, and especially IF3 .
Binding of leaderless mRNAs to the ribosome involves a mechanism that is somewhat different from binding of canonical mRNAs. The binding is dependent on the presence of the initiator tRNA, whereas canonical mRNAs bind independently of the initiator tRNA. Studies with E. coli revealed that the ratio of IF2 to IF3 plays an important role in translation initiation of leaderless mRNAs. Leaderless mRNA is recognized by a 30S-IF2-fMet-tRNAfMet complex equivalent to that formed during translation initiation in eukaryotes (Fig. (Fig.3)3). An increase in the concentration of IF2 enhances the efficiency of leaderless mRNA translation, possibly by recruitment of fMet-tRNAfMet to 30S ribosomal subunits, thus enabling codon-anticodon interaction. Recently, a cell-free translation system was used to show that leaderless mRNAs preferentially interact with 70S ribosomes and are able to proceed from the initiation to the elongation phase even in the absence of initiation factors.
The mRNA wraps around the neck of the 30S ribosomal subunit, with its 5′ end on the platform side and its 3′ end near the shoulder. Structural data for the interaction between mRNA and the ribosome are now available from X-ray crystallographic studies. The new data confirm the general features of the previous models. Interaction between the ASD and SD sequences is located at a large cleft between the head and the back of the platform on the 30S ribosomal subunit (Fig. (Fig.4).4).
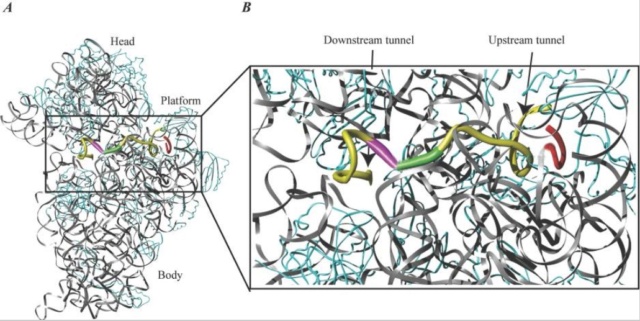
mRNA bound to the 30S ribosomal subunit.
(A) A 36-nucleotide mRNA is bound to the 30S ribosomal subunit. rRNA is shown in grey, mRNA is shown in yellow, and protein is shown in cyan. The ASD sequence of the 16S rRNA is shown in red to indicate the SD-ASD interaction. The P-site initiation codon is shown in green, and the A-site codon is shown in magenta. Note the kink in the mRNA between the two codons.
(B) Close-up of the region indicated in panel A. The upstream and downstream tunnels are marked by arrows. Colors are the same as in panel A. The structure is derived from PDB entry 1JGQ, prepared using the program Ribbons, and rendered in Pov-Ray.
The mRNA wraps around the 30S ribosomal subunit while it passes through the up- and downstream tunnels. A latch-like closure between the head and body on activation of the subunit forms the tunnels. Early studies indicated that binding of mRNA to the ribosome through the SD interaction melts the mRNA secondary structure in the TIR of the mRNA. The mRNA is probably unwound by mRNA helicases before entering the downstream tunnel, since an RNA helix is too large to pass.
The Start Codon Is Chosen
A special tRNA, the initiator tRNA, is charged with methionine and binds to the AUG start codon
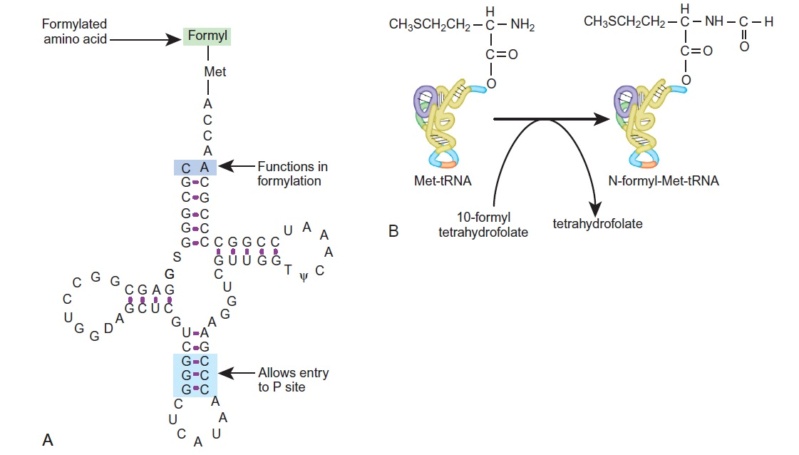
Initiator tRNA Carries N-Formyl-Methionine
(A) The structure of the initiator tRNA, fMet-tRNA, is unique. A CA base pair at the top of the acceptor stem is needed to allow formylation (violet). The initiator tRNA must enter the P-site directly (discussed later), which requires the three GC base pairs in the anticodon stem (blue).
(B) The initiator tRNA is first charged with unmodified methionine. Then a formyl group carried by the tetrahydrofolate cofactor is added to the
methionine.
Selection of the correct AUG in a prokaryotic mRNA requires sequences in the mRNA called Shine-Dalgarno sequences (FIGURE). The ribosome is properly positioned at the correct AUG by base pairing between sequences in the 16S rRNA and the Shine-Dalgarno sequences in the mRNA. Messages with mutated Shine-Dalgarno sequences are poorly translated.
The correct AUG in a eukaryotic mRNA is almost always the first AUG from the 5'-end, but the context of the surrounding sequence is important: GCC(A or G)CCAUGG. The most important features of this sequence are the purine (A or G) 3 bases before the AUG and the G immediately following it (together they influence the efficiency of translation by 10-fold).
In prokaryotes, chemically tagged methionine, N-formyl-methionine (fMet), is attached to the initiator tRNA, whereas in eukaryotes unmodified methionine is used. Consequently, all polypeptide chains begin with methionine, at least when first made. Sometimes the initial methionine (in eukaryotes), or N-formyl-methionine (in prokaryotes), is snipped off later, so mature proteins do not always begin with methionine. In bacteria, even when the fMet is not removed as a whole, the N-terminal formyl group is often removed leaving unmodified methionine at the N-terminus of the polypeptide chain. AUG codons also occur in the middle of messages and result in the incorporation of methionines in the middle of polypeptide chains. So how does the ribosome know which AUG codon to start with? Near the front (the 5' end) of the mRNA of prokaryotes is a special sequence, the ribosome-binding site (RBS), often called the Shine-Dalgarno or S-D sequence, after its two discoverers

Shine-Dalgarno Sequence of mRNA Binds to 16S rRNA
The Shine-Dalgarno sequence on the mRNA is recognized by base pairing with the anti-Shine-Dalgarno sequence on the 16S rRNA. The first AUG downstream of the S-D/anti-S-D site serves as the start codon.
The sequence complementary to this, the anti-Shine-Dalgarno sequence, is found close to the 3' end of the 16S rRNA. Consequently, the mRNA and the 16S rRNA of the ribosome bind together by base pairing between these two sequences. The start codon is the next AUG codon after the ribosome-binding site. Typically, there are about seven bases between the S-D sequence and the start codon. In some cases, the S-D sequence exactly matches the antiS-D sequence and the mRNAs are translated efficiently. In other cases, the match is poorer and translation is less efficient. (Note that eukaryotes do not use an S-D sequence to locate the start of translation; instead, they scan the mRNA starting from the 50-cap) Occasionally, coding sequences even start with GUG (normally encoding valine) instead of AUG. This leads to inefficient initiation and is mostly found for proteins required only in very low amounts, such as regulatory proteins, for example, LacI, the repressor of the lac operon. Note that when GUG acts as the start codon, the same initiator fMettRNA is used as when AUG is the start codon. Consequently, fMet is the first amino acid, even for proteins that start with a GUG codon. This is apparently due to the involvement of the initiation factors, especially IF3.
My comment: Let us suppose there was just a randomly polymerized RNA or DNA polypeptide chain, and the transcription and translation machinery to make proteins had not emerged yet. If the genetic code, and codon-words were not pre-established first, the sequence would be just gibberish, a random sequence. There would not be genetic words (codons), nor syntax being able to be recognized by the translation machinery nor tRNA's, and there would be no assignment of amino acids to make functional proteins.
The Initiation Complex Assembles
Before protein synthesis starts, the two subunits of the ribosome are floating around separately. Because the 16S rRNA, with the anti-Shine-Dalgarno sequence, is in the small subunit of the ribosome, the mRNA binds to a free small subunit. Next the initiator tRNA, carrying fMet, recognizes the AUG start codon. Assembly of this 30S initiation complex needs three proteins (IF1, IF2, and IF3), known as initiation factors, which help arrange all the components correctly. IF2 physically contacts the acceptor stem of the fMet-tRNA, and this interaction is essential to stabilize the initiation complex. IF3 recognizes the start codon and the matching anticodon end of the initiator tRNA. IF3 prevents the 50S subunit from binding prematurely to the small subunit before the correct initiator tRNA is present. Once the 30S initiation complex has been assembled, IF3 departs and the 50S subunit binds. IF1 and IF2 are now released, resulting in the 70S initiation complex.
Proteins known as initiation factors help the ribosomal subunits, mRNA and tRNA assemble correctly.
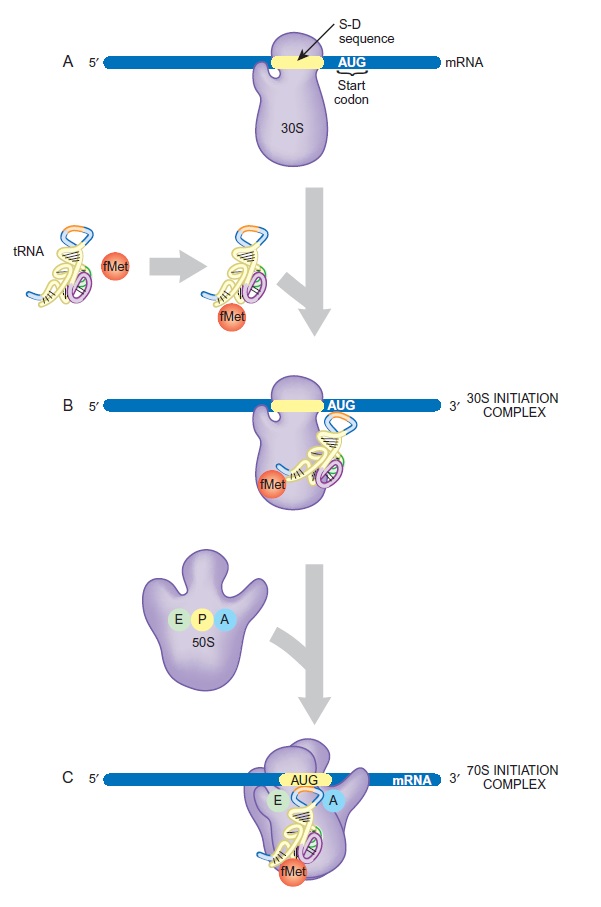
Formation of 30S and 70S Initiation Complexes
(A) The small subunit and the mRNA bind to each other at the Shine-Dalgarno sequence. The start codon, AUG, is just downstream of this site.
(B) The initiator tRNA becomes tagged with fMet and binds to the AUG codon on the mRNA.
(C) The large ribosomal subunit joins the small subunit and accommodates the tRNA at the P-site.
This process consumes energy in the form of GTP, which is split by IF2.
Initiation of Protein Synthesis in eukaryotes
Regulation of cap-dependent translation by eIF4E inhibitory proteins 3 FEBRUARY 2005 1
Eukaryotic messenger RNAs contain a modified guanosine, termed a cap, at their 5' ends. Translation of mRNAs requires the binding of an initiation factor, eIF4E, to the cap structure. Here, we describe a family of proteins that through a shared sequence regulate cap-dependent translation. The biological importance of this translational regulation is immense, and affects such processes as cell growth, development, oncogenic transformation and perhaps even axon pathfinding and memory consolidation.
A single cell never exploits the full panoply of gene products available for its use; at any one time, the transcription of most genes is unwarranted and therefore these genes are silenced. Even for those mRNAs that are synthesized and transported to the cytoplasm, however, there are often other levels of regulation. The past few years have witnessed an explosion in the number of mRNAs whose translation is recognized to be temporally and spatially regulated in various cell types. Although no single mechanism controls the translation of all mRNAs, emerging evidence indicates that the regulated binding of translation initiation factors (eIFs) to the 7-methyl guanosine residue that caps the 50 ends of all nuclear-encoded eukaryotic mRNAs is important. In particular, the interaction of the ribosomal-subunit-associated eIF4G with the cap-bound eIF4E is necessary for cap-dependent translation. A group of factors generically known as eIF4E inhibitory proteins modulate the eIF4G–eIF4E interaction. Whereas some eIF4E inhibitory proteins repress translation by associating with eIF4E on a large number of transcripts, others are tethered ( restrict its movement) to specific subsets of mRNAs through interactions with RNA binding proteins, thus restricting their inhibition of translation to only certain mRNAs. Biologically, the eIF4E inhibitory proteins are enormously important; they control development and cell growth, repress tumour formation, and may influence critical neuronal events such as axon guidance and synaptic plasticity, a phenomenon that may underlie long-term memory storage. Here, we present not only the molecular mechanisms by which some of these proteins control translation, but also describe a few of the biological processes they regulate. Although only a handful of these eIF4E inhibitory proteins have been identified, we suspect that there may be several others that await discovery
The translation initiation machinery
Initiation is the rate-limiting step in translation and is the most common target of translational control. The mRNA 50 cap is bound by eIF4F, a heterotrimeric protein complex that is the focal point for initiation. eIF4G is the backbone of this complex; it interacts not only with eIF4E, but also with eIF4A, an RNA helicase that facilitates ribosome binding and its passage along the 50 untranslated region (UTR) towards the initiation codon. eIF4G also associates with eIF3, a multisubunit factor that bridges the proteins bound to the mRNA’s 50 end with the 40S ribosomal subunit (Fig. 1).
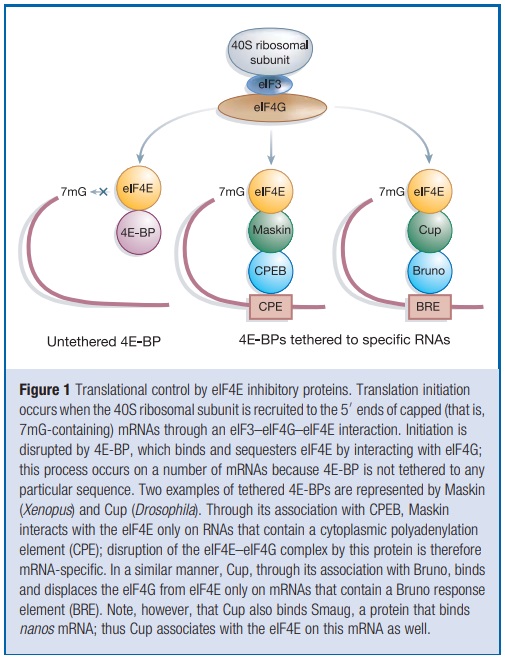
This ribosomal subunit comes ‘pre-charged’ as a ternary complex composed of eIF2, GTP and the initiator methionine-transfer RNA. With the aid of eIF4 initiation factor as well as ATP, this agglomeration of RNA and protein is thought to scan the mRNA in the 50 to 30 direction. When it encounters an AUG start codon in an optimal context, other factors as well as the 60S ribosomal subunit are recruited and polypeptide chain elongation begins . The eIF4E–eIF4G interface is an important target for translational control. The core portion of eIF4G that interacts with eIF4E is small—about 15 amino-acid residues . Strikingly, several other proteins contain similar peptide motifs, and it is this region that competes with eIF4G for binding to eIF4E; in this manner they control the rate of 40S ribosomal subunit association with mRNA, and hence translation initiation. Peptides derived from the regions of eIF4G and an eIF4E inhibitory protein called 4E-BP (for 4E-binding proteins, also known as PHAS-I for phosphorylated heat and acid soluble protein stimulated by insulin) form nearly identical a-helical structures that lie along the same convex region of eIF4E, some distance from this protein’s cap binding site. Peptides with the general sequence YXXXXLf, where f is any hydrophobic amino acid, would probably form similar a-helical structures, implying that other proteins containing this peptide motif could control translation initiation. The original three eIF4E inhibitory proteins, the 4E-BPs, prevent eIF4F complex formation by sequestering available eIF4E. This sequestration results in the inhibition of translation of certain mRNAs that normally require high levels of available eIF4E. The newly discovered eIF4E-binding proteins described below interact with the eIF4E on only specific mRNAs, and do so either because they also interact with certain RNA elements directly, or do so through affiliations with RNA binding proteins.
1.https://www.researchgate.net/profile/Nahum-Sonenberg/publication/8042588_Regulation_of_cap-dependent_translation_by_eIF4E_inhibitory_proteins/links/0912f51467599db728000000/Regulation-of-cap-dependent-translation-by-eIF4E-inhibitory-proteins.pdf
Last edited by Otangelo on Tue Mar 16, 2021 7:36 am; edited 5 times in total