https://reasonandscience.catsboard.com/t1303-challenges-to-endosymbiotic-theory
The origin of eukaryotes is one of the hardest and most intriguing problems in the study of the evolution of life, and arguably, in the whole of biology. On average, the volume of eukaryotic cells is about 15,000 times larger than that of prokaryotic cells. 4 A major problem faced by this scenario (and symbiogenetic scenarios in general) is the mechanistic difficulty of the engulfment of one prokaryotic cell by another. The origin of eukaryotes is a fundamental, forbidding evolutionary puzzle. The scenario of eukaryogenesis, and in particular the relationship between endosymbiosis and the origin of eukaryotes, is far from being clear. Compared to archaea and bacteria (collectively, prokaryotes), eukaryotic cells are three to four orders of magnitude larger in volume and display a qualitatively higher level of complexity of intracellular organization. Eukaryotic cells function on different physical principles compared to prokaryotic cells, which is directly due to their (comparatively) enormous size. The gulf between the cellular organizations of eukaryotes and prokaryotes is all the more striking because no intermediates have been found. So intimidating is the challenge of eukaryogenesis that the infamous notion of irreducible complexity’ has sneaked into serious scientific debate 2 . The diversity of the outcomes of phylogenetic analysis, with the origin of eukaryotes scattered around the archaeal diversity, has led to considerable frustration and suggested that a ‘phylogenomic impasse’ has been reached, owing to the inadequacy of the available phylogenetic methods for disambiguating deep relationships. The evolutionary trajectory of modern eukaryotes is distinct from that of prokaryotes. Data from many sources give no direct evidence that eukaryotes evolved by genome fusion between archaea and bacteria. Nuclei, nucleoli, Golgi apparatus, centrioles, and endoplasmic reticulum are examples of cellular signature structures (CSSs) that distinguish eukaryote cells from archaea and bacteria.
Christopher S. Winefield: Evolutionary Analysis of Aspartate Aminotransferases : 22 June 1994
It is widely believed that mitochondria are derived from an endosymbiotic organism that colonized the ancestral eukaryote and that chloroplasts arose from a similar, but the distinct, event (Gray 1989). At the time of colonization, a complete but distinct alternative genome was present in the protomitochondria (and protochloroplast). The mitochondrial genome is now compressed and codes for only a few of the genes that are necessary for, and specific to, mitochondrial function--the balance of these genes now being present within the nuclear genome. This tendency to condense the mitochondrial genome is less marked for green plants where the mitochondrial genome is typically 200 kb, at an intermediate stage in the yeast mitochondrial genome (78 kb), and most pronounced in vertebrate mitochondria where the genome is about 16kb. There are at least two possible mechanisms that could account for the transfer of genetic information from the mitochondrial to nuclear genomes. In one scheme, it is proposed that genes may have duplicated in the nuclear genome and mutated until they were able to take over the role of the mitochondrial genes and subsequently the mitochondrial genes were lost. An alternative proposal is that the genes themselves were "copied" or transferred from the mitochondrial to the nuclear genome and were then deleted from the mitochondria. These mechanisms are not mutually exclusive and it is possible that both mechanisms have operated to produce the current observed state. A further mechanism that may account for different forms of an enzyme being present in the mitochondria and cytosol is differential splicing of a precursor mRNA. Such a mechanism is apparently responsible for the mitochondrial and cytosolic forms of fumarase found in vertebrates
https://pubmed.ncbi.nlm.nih.gov/7769621/
Intracellular Calcium, page 578
Most biology textbooks now tell us that organelles such as chloroplasts and mitochondria, both of which have circular DNA, evolved through endosymbiosis – a hypothesis promoted by Lynn Margulis (Margulis, 1991). However, the evidence for this is weak, and a much more likely origin of organelles such as the ER, mitochondria, lysosomes, peroxisomes, secretory vesicles, chloroplasts and the tonoplast in plants is invagination of membranes. Comparisons of mitochondrial DNA throughout animals, plants and eukaryotic microbes supports the hypothesis that mitochondria arose only once in evolution, and were from a proto-bacterial cell (Lang et al., 1999). But, the genomes of mitochondria and chloroplasts are too small to code for the genes necessary for a complete organism.
Human mitochondrial DNA has just 16 569 base pairs, coding for only 37 genes, which are all essential for mitochondrial function, but far too little for a cell to survive. Thirteen of these genes produce proteins essential for ATP synthesis by oxidative phosphorylation, the other 25 coding for tRNA and rRNA, necessary for mitochondrial protein synthesis. Mitochondrial ribosomes are like bacterial ribosomes. None are involved in Ca2+ signaling. Yet, E. coli has some 300 essential genes which cannot be knocked-out without killing the cell, yet there are some 1500 proteins found inside a mitochondrion, several of which are involved in transporting Ca2+ in and out or responding to a rise in intra-mitochondrial free Ca2+. Mitochondrial divide, make proteins, make ATP, and carry out several other biochemical pathways, such as fatty acid oxidation. So if mitochondria originated from an endosymbiont such as Rickettsia there are three problems:
1. How did the endocytosed bacterium survive and multiply if its internal environment was oxidizing? The cytosol of all cells is reducing, preventing the formation of S–S bonds and damaging oxidative reactions involving reactive oxygen species. But, remember the first eukaryotes formed before there was significant oxygen in the atmosphere. So oxidative phosphorylation in mitochondria must have evolved after photosynthesis, some 2000 million years ago.
2. Since cells need at least 1200 proteins to survive, replicate, and synthesize their own building blocks, what happened to the proteins essential for nucleotide and nucleic acid, and protein synthesis, and the reactions necessary for ATP synthesis, e.g. glycolysis?
3. How did the 1500 or so mitochondrial proteins in the main genome become targeted to the mitochondria, if they were lost by the initial endosymbiont?
For the endosymbiotic hypothesis to work, the primitive bacterium engulfed by the eukaryote precursor must have lost over 90% of its genes, these being taken up by the nuclear genome. Furthermore, most of the proteins involved in Ca2+ signaling must have come from another source.
Plastid DNA contains just 60–100 genes, whereas a typical cyanobacterium DNA codes for 1500. The rest of the chloroplast proteins, like mitochondria, are coded for by the nuclear genome. A chloroplast genome of around 140 kb is comparable to a large bacteriophage, such as T4 whose genome is about 65 kb.
The start of the sequence for the origin of a mitochondrion or chloroplast would have to be a bacterium or cyanobacterium being taken up by the eukaryotic precursor. Then this has to lose genes, which are captured by the main genome. Then some of these genes have to have targeting sequences added them and they have to change several codons so that their genetic code matches the main genome, and not the proteins that remain in the mitochondrial or chloroplast genome, since there are differences in the genetic code between non-plant mitochondria and nuclear DNA, and losses of anticodons in chloroplasts. This is too many ‘ifs’ and ‘buts’ to be credible!
Did eukaryotes evolve from prokaryotic cells?
https://reasonandscience.catsboard.com/t1568-did-eukaryotes-evolve-from-prokaryotic-cells
1. There are no true intermediates in the prokaryote-to-eukaryote transition. More than 20 different versions of endosymbiotic theory have been presented in the literature to explain the origin of eukaryotes and their mitochondria. The origin of eukaryotes is certainly one of early evolution's most important topics. “Throughout 150 years of the science of bacteriology, there is no evidence that one species of bacteria has changed into another... Since there is no evidence for species changes between the simplest forms of unicellular life, it is not surprising that there is no evidence for evolution from prokaryotic [i.e., bacterial] to eukaryotic [i.e., plant and animal] cells, let alone throughout the whole array of higher multicellular organisms.” The organizational complexity of the eukaryotes is so much greater than that of the prokaryotes that it is difficult to visualize how a eukaryote could have arisen from any known prokaryote (Hickman et al., 1997, p. 39). In eukaryotes the mitochondria produce most of the cell’s ATP (anaerobic glycolysis also produces some) and in plants the chloroplasts can also service this function. The mitochondria produce ATP in their internal membrane system called the cristae. Since bacteria lack mitochondria, as well as an internal membrane system, they must produce ATP in their cell membrane which they do by two basic steps. The bacterial cell membrane contains a unique structure designed to produce ATP and no comparable structure has been found in any eukaryotic cell (Jensen, Wright, and Robinson, 1997).
2. The Darwinian Basis of the Prokaryote-to-Eukaryote Transition Collapses
https://reasonandscience.catsboard.com/t1568-did-eukaryotes-evolve-from-prokaryotic-cells#3782
Mitochondria has a different genetic code, and there is no viable route for the evolution of the genetic code. Mitochondria use a slight variation on the conventional genetic code (for example, the codon UGA is a stop codon in the conventional code, but encodes for Tryptophan in mitochondria). This implicates that the genes of the ingested prokaryotes would need to have been recoded on their way to the nucleus. The situation becomes even worse when one considers that, in eukaryotic cells, a mitochondrial protein is coded with an extra length of polypeptide which acts as a "tag" to ensure that the relevant protein is recognised as being mitochondrial and dispatched accordingly. The significant number of specific co-ordinated modifications which would be required to facilitate such a transition, therefore, arguably make it exhibitive of irreducible complexity.
The different genetic codes
https://reasonandscience.catsboard.com/t2277-the-different-genetic-codeses
The National Center for Biotechnology Information (NCBI), currently acknowledges nineteen different coding languages for DNA. And i list 31 different ones.
If the mitochondria in invertebrates use a different genetic code from the mitochondria in vertebrates, and both of those codes are different from the “universal” genetic code, what does that tell us? It means that the eukaryotic cells that eventually evolved into invertebrates must have formed when a cell that used the “universal” code engulfed a cell that used a different code. However, the eukaryotic cells that eventually evolved into vertebrates must have formed when a cell that used the “universal” code engulfed a cell that used yet another different code. As a result, invertebrates must have evolved from one line of eukaryotic cells, while vertebrates must have evolved from a completely separate line of eukaryotic cells. But this isn’t possible, since evolution depends on vertebrates evolving from invertebrates. Now, of course, this serious problem can be solved by assuming that while invertebrates evolved into vertebrates, their mitochondria also evolved to use a different genetic code. However, I am not really sure how that would be possible. After all, the invertebrates spent millions of years evolving, and through all those years, their mitochondrial DNA was set up based on one code. How could the code change without destroying the function of the mitochondria? At minimum, this adds another task to the long, long list of unfinished tasks necessary to explain how evolution could possibly work. Along with explaining how nuclear DNA can evolve to produce the new structures needed to change invertebrates into vertebrates, proponents of evolution must also explain how, at the same time, mitochondria can evolve to use a different genetic code!
3. Membranes of dauther cells are only inherited by membranes of mother cells through fission.
Intracellular Compartments and Protein Sorting
https://reasonandscience.catsboard.com/t3048-intracellular-compartments-and-protein-sorting
Unlike a bacterium, which generally consists of a single intracellular compartment surrounded by a plasma membrane, a eukaryotic cell is elaborately subdivided into functionally distinct, membrane-enclosed compartments. Each compartment, or organelle, contains its own characteristic set of enzymes and other specialized molecules, and complex distribution systems transport-specific products from one compartment to another. To understand the eukaryotic cell, it is essential to know how the cell creates and maintains these compartments, what occurs in each of them, and how molecules move between them. Proteins confer upon each compartment its characteristic structural and functional properties. They catalyze the reactions that occur there and selectively transport small molecules into and out of the compartment. For membrane-enclosed organelles in the cytoplasm, proteins also serve as organelle-specific surface markers that direct new deliveries of proteins and lipids to the appropriate organelle. An animal cell contains about 10 billion (10^10) protein molecules of perhaps 10,000 kinds, and the synthesis of almost all of them begins in the cytosol, the space of the cytoplasm outside the membrane-enclosed organelles. Each newly synthesized protein is then delivered specifically to the organelle that requires it. . By tracing the protein traffic from one compartment to another, one can begin to make sense of the otherwise bewildering maze of intracellular membranes
4. Mitochondrial membrane biogenesis: phospholipids and proteins go hand in hand 1
https://reasonandscience.catsboard.com/t2128-cell-membranes-origins-through-natural-mechanisms-or-design
Mitochondrial membrane biogenesis requires the import and synthesis of proteins as well as phospholipids.The biochemical approach of Kutik et al. (2008) uncovered an unexpected role of the mitochondrial translocator assembly and maintenance protein, Tam41, in the biosynthesis of cardiolipin (CL), the signature phospholipid of mitochondria. The genetic analyses of Osman et al. (2009) led to the discovery of a new class of mitochondrial proteins that coordinately regulate CL and phosphatidylethanolamine, another key mitochondrial phospholipid. These elegant studies highlight overlapping functions and interdependent roles of mitochondrial phospholipid biosynthesis and protein import and assembly
5. The mitochondrial inner membrane has a unique composition of proteins and phospholipids, whose interdependence is crucial for mitochondrial function.
6. Most Organelles Cannot Be Constructed De Novo: They Require Information in the Organelle Itself
https://reasonandscience.catsboard.com/t2122-most-organelles-cannot-be-constructed-de-novo-they-require-information-in-the-organelle-itself
https://reasonandscience.catsboard.com/t2397-the-interdependency-of-lipid-membranes-and-membrane-proteins
A cell cannot produce the cell membrane de novo from scratch. It inherits it. Daughter cell membranes come only from mother cell membranes.
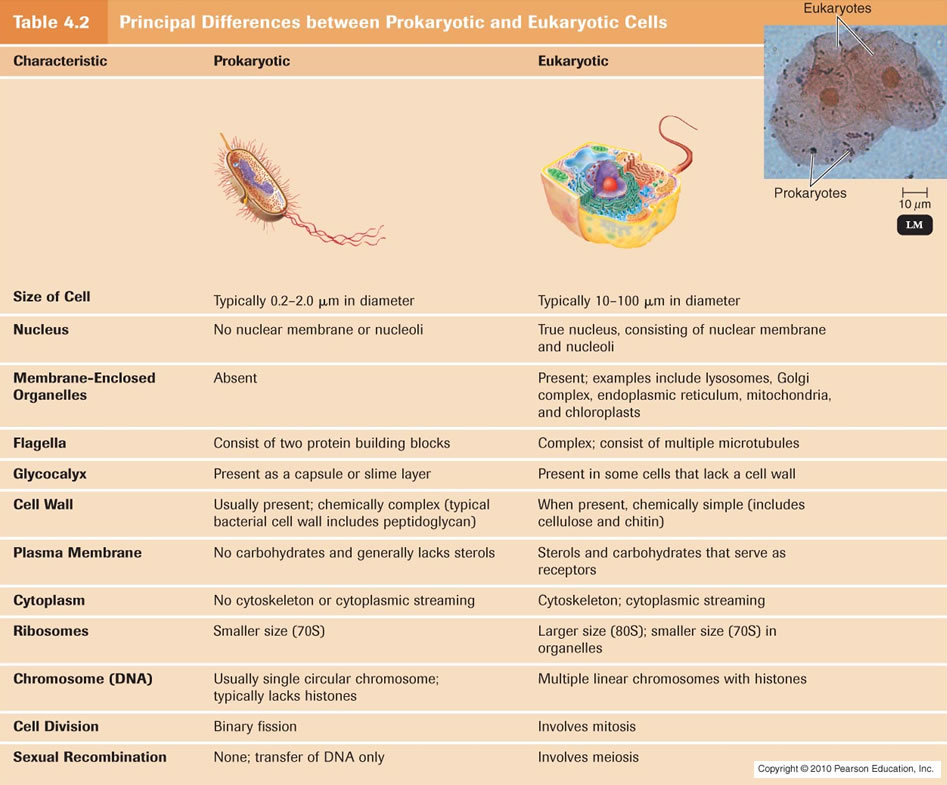
Functional Anatomy Of Prokaryotic And Eukaryotic Cells
With regret, ENV recently noted the passing of biologist Lynn Margulis. Margulis, a scientist whom I admired greatly, was never a stranger to controversy, going so far as to call neo-Darwinism "a complete funk" and asserting that "The critics, including the creationist critics, are right about their criticism. It's just that they've got nothing to offer by intelligent design or 'God did it.' They have no alternatives that are scientific." She was a scientist who wasn't afraid to think creatively, disregarding the scorn of her colleagues. According to the Telegraph, a response to one grant application she made said: "Your research is crap. Don't ever bother to apply again."
Lynn Margulis took a controversial view on how evolution works, stressing the importance of symbiotic and co-operative relationships over competition. This concept of evolution inspired what is now recognized as her most notable idea, the notion that the eukaryotic mitochondrion -- the power plant of the cell -- was acquired by virtue of an endosymbiotic event. Endosymbiotic theory essentially maintains that mitochondria arose by virtue of a symbiotic union of prokaryote cells. The nearest living relative to the mitochondrion is thought to be the alpha-proteobacteria Rickettsia (Emelyanov, 2000; Andersson et al., 1998). Chloroplasts are also thought to have arisen in a similar manner from the photosynthetic cyanobacteria.
In November 2010, I drew attention to a paper in Nature by Nick Lane and Bill Martin, who showed that the prokaryote-to-eukaryote transition was effectively impossible without the energy demands, pertinent to the biggest event of gene manufacture in the history of life on earth, being met by the mitochondrial processes of oxidative phosphorylation and the electron transport chain. The bacterial cell alone could not meet these energy demands. The evidence that is typically offered for endosymbiotic theory includes the following:
Mitochondria possess a circular genome (lacking in introns and independent from the nuclear DNA) in which transcription is coupled to translation, characteristic of bacterial DNA. There are also some other notable similarities. For example, in both mitochondria and Mycoplasma, the codon UGA specifies the amino acid Tryptophan , whereas in the conventional code it serves as a stop codon. Mitochondria divide and replicate independently of host cell division and do so in a manner akin to binary fission, possessing homologues of the bacterial division protein FtsZ .They are enclosed by a double-membrane.Mitochondria and bacteria are of a similar size and shape.Circular Mitochondrial Genome. As noted, one of the core arguments for endosymbiosis points to its circular genome. What is often not noted, however, are the cases where eukaryotic mitochondria have linear genomes with eukaryotic telomeres. Indeed, two strains of the same species of yeast differ with respect to the linearity or circularity of their mitochondrial genome .In the case of linear chromosomes, the DNA polymerase enzymes are unable to replicate right to the end of the chromosome. This is because the enzymes are unable to replace the lagging strand's terminal RNA primer. Unless there is a mechanism for circumventing this, it will result in the chromosomes shortening after each round of replication (in eukaryotes, the enzyme telomerase attaches extra DNA to the chromosomal ends).
This means that the transition from genome circularity to linearity -- a fete in itself given the changes that have to be made to the mode of replication -- must happen in concert with the evolution of a mechanism to prevent progressive chromosomal shortening.
In order to have a transition from prokaryotic to eukaryotic dna replication, telomerase enzymes must arise simultaniously, to prevent the shortening of the telomere region after every replication.
Telomerase, also called telomere terminal transferase,[1] is a ribonucleoprotein that adds the polynucleotide "TTAGGG" to the 3' end of telomeres, which are found at the ends of eukaryotic chromosomes. A telomere is a region of repetitive sequences at each end of a chromatid, which protects the end of the chromosome from deterioration or from fusion with neighbouring chromosomes. 5
Such an evolutionary transition is far from trivial. Biologist Albert de Roos writes,n linear mitochondrial chromosomes various different mechanisms to "prevent" shortening exist, ranging from hairpin loops and self-priming to protein-assisted primer synthesis (see here). The telomeric regions of mitochondrial chromosomes do not seem to have a direct phylogenetic relation since they use other proteins and mechanisms than nuclear telomeres. Thus, it is difficult to deduce evolutionary pathways purely based on phylogenetic data on telomeres and mechanisms for end replication.Furthermore, mitochondrial genes often do possess introns . These are particularly prevalent in the mtDNA of fungi and plants. The mitochondrial genetic code may also be slightly different from that of bacteria .Mitochondrial DNA Replication:
The claim one often hears is that circular mitochondrial DNA replication resembles bacterial binary fission. While this is true, in at least some respects, there are also important differences. For example, many of the key components are of eukaryotic origin and replication beginning at the Displacement (D-) loop (Fish et al., 2004; Clayton, 1996) is not the same as bacterial DNA replication.
Double Membrane
It is frequently asserted that the double membrane of mitochondria provides evidence for its endosymbiotic origin. There are, however, important differences between bacterial and mitochondrial membranes. Albert de Roos observes,
The Size and Shape of MitochondriaThe bacterial membrane is one of the basic characteristics that distinguish bacteria from eukaryotes, see some examples here. In order for mitochondria to resemble bacterial membranes, they should share characteristics such as a cell wall with peptidoglycan and lipopolysaccharides, gram-staining and antibiotic sensitivity. Some effects of antibiotics have been seen with both bacteria and mitochondria, but the effect is minor while the use of antibiotics is based on the principle that they distinguish between bacteria and eukarytes, including the mitochondrion (here). Until then, the selection of a few apparent similarities while ignoring the many differences does not indicate a bacterial origin for mitochondria. On the contrary, the fact that their membranes are so different as well as the fact that nearly all genes are encoded by the nucleus is primarily evidence against a bacterial origin.
Even though some shared characteristics may be found, we have to realize that bacterial and eukaryotic membranes are fundamentally different. It seems virtually impossible to change all fundamental bacterial membrane characteristics and replace them with a eukaryotic counterpart without loosing membrane integrity. The differences between the membranes of mitochondria and the cell walls of bacteria make the endosymbiotic theory mechanistically difficult. It seems quite clear that bacterial membranes do not change easily into other membranes, and frankly I don't see any scenarios in which to change all these membrane components without drastically affecting fitness.
The argument based on the size and shape of mitochondria is one that has been turned on its head in recent years, being transformed from an argument forendosymbiosis to one against it. These organelles are now acknowledged in the literature to be better understood as dynamic reticular structures (see this linkfor references).
Electron micrographs displaying cross-sections of mitochondria portrayed the mitochondrion as a sphere. However, when one looks at 3D models of the organelle, the reality is somewhat different. You can take a look at some of these images by going here, here, here, or here.
The Lack of a Mechanism
By far the most potent challenge to the endosymbiotic origin of eukaryotic mitochondria is the lack of a viable mechanism, perhaps most particularly with respect to the transfer of genes from the mitochondrion to the nucleus.
For one thing, there are the variants on the conventional genetic code. This means that, over the course of their transfer to the nucleus, the genes would need to be "recoded" so as to comply with the conventional genetic code. For example, recognizing UGA as a stop codon instead of the codon for Tryptophan(or vice versa) would cause cellular mayhem.
Secondly, mitochondrial proteins made at the ribosomes in the eukaryote cytoplasm need to be identified as such to ensure that they are properly dispatched (this is normally done by attaching a "label" in the form of an extra length of polypeptide to the protein). This would require a coincidental modification of the correct structural gene (which seems unlikely). Biologist Timothy G. Standish [url=http://www.google.co.uk/url?sa=t&rct=j&q=if genes were to move from the mitochondria to the nucleus they would have to somehow pick up the leader sequences necessary to signal for transport before they could be]notes[/url],
Albert de Roos explains,
- If genes were to move from the mitochondria to the nucleus they would have to somehow pick up the leader sequences necessary to signal for transport before they could be functional.
- While leader sequences seem to have meaningful portions on them, according to Lewin (1997, p251) sequence homology between different sequences is not evident, thus there could be no standard sequence that was tacked on as genes were moved from mitochondria to nucleus.
- Alternatively, if genes for mitochondrial proteins existed in the nucleus prior to loss of genes in the mitochondria, the problem remains, where did the signal sequences come from? And where did the mechanism to move proteins with signal sequences on them come from?
All evolutionary theories must offer an explanation in mechanistic terms of how it should or could have happened in order to be tested. The difficult thing with the endosymbiotic theory is that it proposes no real mechanism and most textbooks show the simplistic picture of a cell that swallows another cell that becomes a mitochondrion. Unfortunately, it is not so simple as that. There is a difference between the process of endosymbiosis and its incorporation in the germline, necessitating genetic changes. What were those changes? What was the host? Was it a fusion, was it engulfment, how did the mitochondrion get its second membrane, how did two genomes in one cell integrate and coordinate? The theory is also strongly teleological, illustrated by the widely used term 'enslavement'. But how do you enslave another cell, how do you replace its proteins and genes without affecting existing functions? The existence of obligate bacterial endosymbionts in some present eukaryotes is often presented as a substitute for a mechanism, but they remain bacteria and give not rise to new organelles. So, before we can speak of the endosymbiotic as a testable scientific theory, we need a mechanistic scenario which is lacking at the moment.
When we do try to envision a mechanistic scenario based on the endosymbiotic theory, we quickly run into problems. Genetic mutations that allow bacteria to thrive in the cytoplasm would not be strategic for survival. Anaerobic cells normally do not survive in environment that contains oxygen, while the endosymbiont would need oxygen in order to present fitness advantage. The two organisms would initially compete for energy sources since bacteria are users of ATP and do not export it. The extensive gene transfer that is needed in the endosymbiotic theory would wreak havoc in a complex genome since frequent insertion of random pieces of mitochondrial DNA would disrupt existing functions. Furthermore, gene transfer is a multi-step process were genes need to be moved to the nucleus, the different genetic code of mitochondria needs to be circumvented, the genes need to be expressed correctly, as well as imported back into the mitochondria in order to be functional. All in all, mechanistic scenarios for the endosymbiotic theory imply many non-functional intermediates or would just be plain harmful to an organism. Therefore, the endosymbiotic theory is in contrast with the concept of gradualism that forms the basis of modern evolutionary theory.
Furthermore, this gene transfer must have taken place at a time extremely early in the history of eukaryotes, substantially reducing the window of time in which gene transfer could have occurred.
Summary and Conclusion
While we find examples of similarity between eukaryotic mitochondria and bacterial cells, other cases also reveal stark differences. In addition, the sheer lack of a mechanistic basis for mitochondrial endosymbiotic assimilation ought to -- at the very least -- give us reason for caution and the expectation of some fairly spectacular evidence for the claim being made. At present, however, such evidence does not exist -- and justifiably gives one cause for skepticism.
Graham JonesSaturday:
I read Nick Lanes's The Vital Question, and there's something that puzzles me, which relates to this article: The energetics of genome complexity, Nick Lane & William Martin, 2010. (http://www.nature.com/nature/journal/v467/n7318/abs/nature09486.html)
They do a back-of-the-envelope calculation. Prokaryotes metabolise about 3x faster than eukaryotes in terms of watts/g, but eukaryotic cells are 15,000x bigger, so eukaryotic cells have 5000x times as much power in terms of watts/cell. Eukaryotes have 4x as many genes as prokaryotes, so they have 1200x as much power per gene.
About 80% of a cell's power is used for making proteins, so it seems eukaryotes make about 1200x as much protein per gene. I started wondering about the mechanics of that. Transcription rates and translation rates per nucleotide are at least as fast in prokaryotes, and there's no introns to transcribe or splice out, so you'd think prokaryotes would make more protein per gene, not 1200x less.
Eukaryotic cells cells live longer of course, and most are diploid, so have two copies of each gene. Some are polyploid or multinucleated - and I think Lane and Martin's "15,000x bigger" includes some eukaryotic cells with lots of nuclei. A fair comparison would be of (rate of protein synthesis)/(gene copy).
So I got some numbers, from the book Cell Biology by the Numbers, and the biomumbers site (http://bionumbers.hms.harvard.edu/). I found it easiest to get figures for cell volumes. I assume (protein weight)/volume is similar for these cells. They are for organisms when they are growing fast (but not as fast as possible). V = volume in um^3 per gene copy per hour.
E Coli: volume 0.7 um^3, #gene copies 4400, doubling time 1h. V = 1.6e-4
Budding yeast: volume 37 um^3, #gene copies 6000, doubling time 3h. V = 20e-4
C Elegans: volume 1800 um^3, #gene copies 40000, doubling time 10h. V = 45e-4
Euglena gracilis: volume 3700 um^3, #gene copies 60000, doubling time 12h. V = 50e-4
It seems that these eukaryotic cells make protein 10-30x faster than E Coli, per gene copy. Looks very odd to me. Presumably the limiting factor for prokaryotes is power, as Lane and Martin suggest. Apparently prokaryotes have the molecular machinery to make protien faster than eukaryotes, but they hardly ever have enough energy to run the machinery anywhere near full speed.

A simplified view of the endosymbiosis theory.
(a) According to this concept, modern mitochondria were derived from purple bacteria, also called α-proteobacteria. Over the course of evolution, their characteristics changed into those found in mitochondria today.
(b) A similar phenomenon occurred for chloroplasts, which were derived from cyanobacteria (blue-green bacteria), a bacterium that is capable of photosynthesis.
Because mitochondria look like prokaryotes, it was assumed that eukaryotic cells came into existence when one prokaryote swallowed another prokaryote. A problem with this idea is that prokaryotic cells lack the ability to swallow other cells. Eukaryotic cells can, but every eukaryote we know about has mitochondria. It's not clear which came first: the ability to swallow other cells, or the mitochondria.
Examples of ecological circumstances driving genome reduction are seen in many intracellular endosymbionts and parasites, which gain few genes but lose many genes responsible formetabolic flexibility The mitochondrion is even more extreme in its reductive evolution; its ancestral bacterial genome has been reduced to a vestigial microgenome supported by a predominantly eukaryote proteome. Genomes of modern mitochondria encode between 3 and 67 proteins, whereas the smallest known free-living a-proteobacterium (Bartonella quintana) encodes 1100 proteins. Taking Bartonella as a minimal genome for the freeliving ancestor of mitochondria, nearly all of the bacterial coding sequences have been lost from the organelle, though not necessarily from the eukaryote cell. The mitochondrial genome of the protist Reclinomonas americana is the largest known but has still lost more than 95% of its original coding capacity. This abbreviated account of genome reduction illustrates the Darwinian view of evolution as a reversible process in the sense that ‘‘eyes can be acquired and eyes can be lost.’’ Genome evolution is a two-way street. This bidirectional sense of reversibility is important as an alternative to the view of evolution as a rigidly monotonic progression from simple to more complex states, a view with roots in the 18th-century theory of orthogenesis. Unfortunately, such a model has been tacitly favored by molecular biologists who appeared to view evolution as an irreversible march from simple prokaryotes to complex eukaryotes, from unicellular to multicellular. The many well documented instances of genome reduction provide a necessary corrective measure to the often-unstated assumption that eukaryotes must have originated from prokaryotes.
1) https://en.wikipedia.org/wiki/Pre-replication_complex
2) http://nar.oxfordjournals.org/content/27/17/3389.full
3) http://www.answersingenesis.org/articles/2006/10/11/endosymbiotic-theory
4) http://www.evolutionnews.org/2012/01/on_the_origin_o054891.html
5) https://en.wikipedia.org/wiki/Telomerase
6) Intracellular Calcium, page 578
Last edited by Otangelo on Fri Aug 25, 2023 6:56 pm; edited 31 times in total



