The physiology and habitat of the last universal common ancestor
https://reasonandscience.catsboard.com/t2980-the-physiology-and-habitat-of-the-last-universal-common-ancestor
Evolution has been tested and it fails. 60,000 generations of bacteria, and all they got, are bacteria. No hint of a transition zone to a new organismal limb or improvement of complexity. Fail.
25 JULY 2016
The concept of a last universal common ancestor of all cells (LUCA, or the progenote) is central to the study of early evolution and life’s origin, yet information about how and where LUCA lived is lacking. Because these proteins are not universally distributed, they can shed light on LUCA’s physiology. Their functions, properties and prosthetic groups depict LUCA as
anaerobic, CO2-fixing, H2-dependent with a Wood–Ljungdahl pathway, N2-fixing and thermophilic.
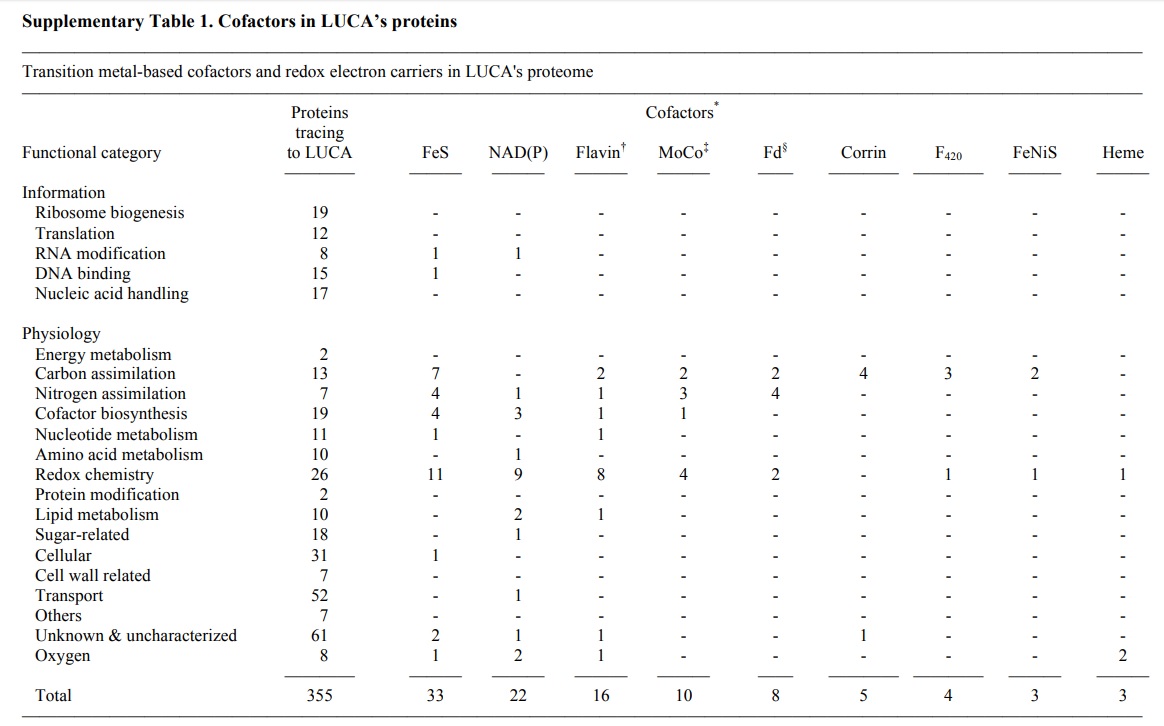
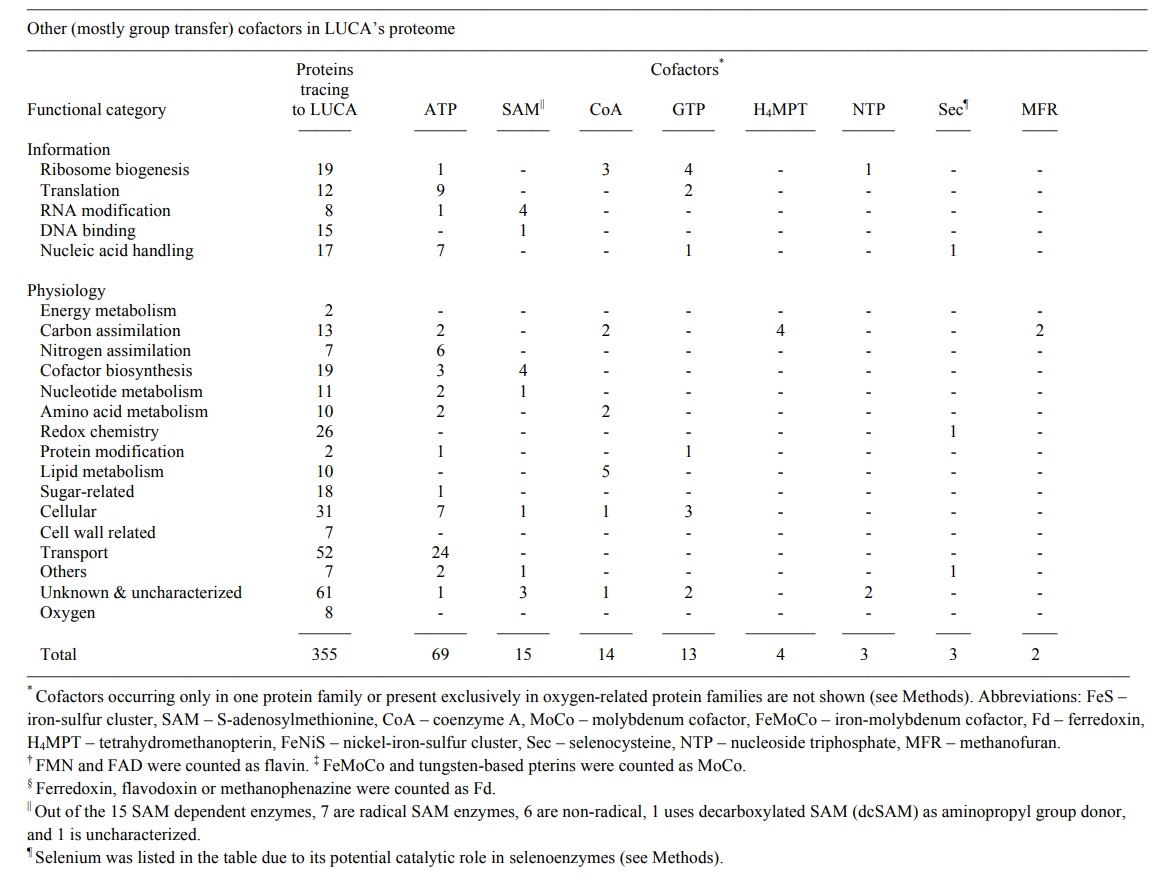
The list with 355 protein annotations is provided by supplementary table 2 near the end of the page:
https://www.nature.com/articles/nmicrobiol2016116
Ribosome biogenesis
Translation
RNA modification
DNA binding
Nucleic acid handling
Energy metabolism
Carbon assimilation
Nitrogen assimilation
Cofactor biosynthesis
Nucleotide metabolism
Amino acid metabolism
Redox chemistry
Protein modification
Lipid metabolism
Sugar-related
Cellular
Cell wall-related
Transport
Others
Unknown & uncharacterized & predicted
Oxygen
The cell is irreducibly complex
https://reasonandscience.catsboard.com/t1299-abiogenesis-the-cell-is-irreducibly-complex
Biological Cells are irreducibly complex:
1. Cell subunits and compartments form a complex system that is useful only in the completion of a much larger system that is able to keep the basic functions of life. A minimal Cell, in order to permit life, according to an article in Science magazine, from 2016, requires a minimal genome of about 473gene products, and at least 438 proteins, each fully set up and functional for specific tasks. A discrete minimal size of each individual protein complex formed by multiple subunits and cofactors is required in order to be functional. And it only operates when interconnected and in a joint venture similar to a robot in a production line, and precise energy supply.
2. Cells must be created and be functional, all at once. As Graham Cairns-Smith noted, this system has to be fixed in its essentials through the critical interdependence of subsystems. Irreducibly complex and interdepend systems cannot evolve but depend on intelligence with foreknowledge on how to build discrete parts with distant goals.
3. Therefore, intelligent design is the best explanation of the origin of living, self-replicating cells.
Chemistry and the Missing Era of Evolution: A. Graham Cairns-Smith
We can see that at the time of the common ancestor, this system must already have been fixed in its essentials, probably through a critical interdependence of subsystems. (Roughly speaking in a domain in which everything has come to depend on everything else nothing can be easily changed, and our central biochemistry is very much like that.
https://sci-hub.tw/https://www.ncbi.nlm.nih.gov/pubmed/18260066
The role of natural selection in the origin of life
Unlike living systems that are products of and participants in evolution, these prebiotic chemical structures were not products of evolution. Not being yet intricately organized, they could have emerged as a result of ordinary physical and chemical processes.
https://www.ncbi.nlm.nih.gov/pubmed/20407927
chemist Wilhelm Huck, professor at Radboud University Nijmegen
A working cell is more than the sum of its parts. "A functioning cell must be entirely correct at once, in all its complexity
http://www.ru.nl/english/@893712/protocells-formed/
https://www.sciencedaily.com/releases/2013/07/130702100115.htm
How Structure Arose in the Primordial Soup
About 4 billion years ago, molecules began to make copies of themselves, an event that marked the beginning of life on Earth. A few hundred million years later, primitive organisms began to split into the different branches that make up the tree of life. In between those two seminal events, some of the greatest innovations in existence emerged: the cell, the genetic code and an energy system to fuel it all. ALL THREE of these are ESSENTIAL to life as we know it, yet scientists know disappointingly little about how any of these remarkable biological innovations came about.
https://www.scientificamerican.com/article/how-structure-arose-in-the-primordial-soup/
We investigated all clusters and phylogenetic trees for 6.1 million protein-coding genes from sequenced prokaryotic genomes in order to reconstruct the microbial ecology of LUCA. Among 286,514 protein clusters, we identified 355 protein families (∼0.1%) that trace to LUCA by phylogenetic criteria.
LUCA’s biochemistry was replete with FeS clusters and radical reaction mechanisms. Its cofactors reveal dependence upon transition metals, flavins, S-adenosyl methionine, coenzyme A, ferredoxin, molybdopterin, corrins and selenium. Its genetic code required nucleoside modifications and S-adenosyl methionine-dependent methylations. The 355 phylogenies identify clostridia and methanogens, whose modern lifestyles resemble that of LUCA, as basal among their respective domains. LUCA inhabited a geochemically active environment rich in H2, CO2 and iron. The data support the theory of an autotrophic origin of life involving the Wood–Ljungdahl pathway in a hydrothermal setting.
The last universal common ancestor (LUCA) is an inferred evolutionary intermediate that links the abiotic phase of Earth’s history with the first traces of microbial life in rocks that are 3.8–3.5 billion years of age . Although LUCA was long considered the common ancestor of bacteria, archaea and eukaryotes newer two-domain trees of life have eukaryotes arising from prokaryotes making LUCA the common ancestor of bacteria and archaea. Previous genomic investigations of LUCA’s gene content have focused on genes that are universally present across genomes, LUCA had 30–100 proteins for ribosomes and translation. To identify genes that can illuminate the biology of LUCA, we took a phylogenetic approach. By focusing on the phylogeny rather than universal gene presence, we can identify genes involved in LUCA’s physiology—the ways that cells access carbon, energy and nutrients from the environment for growth.
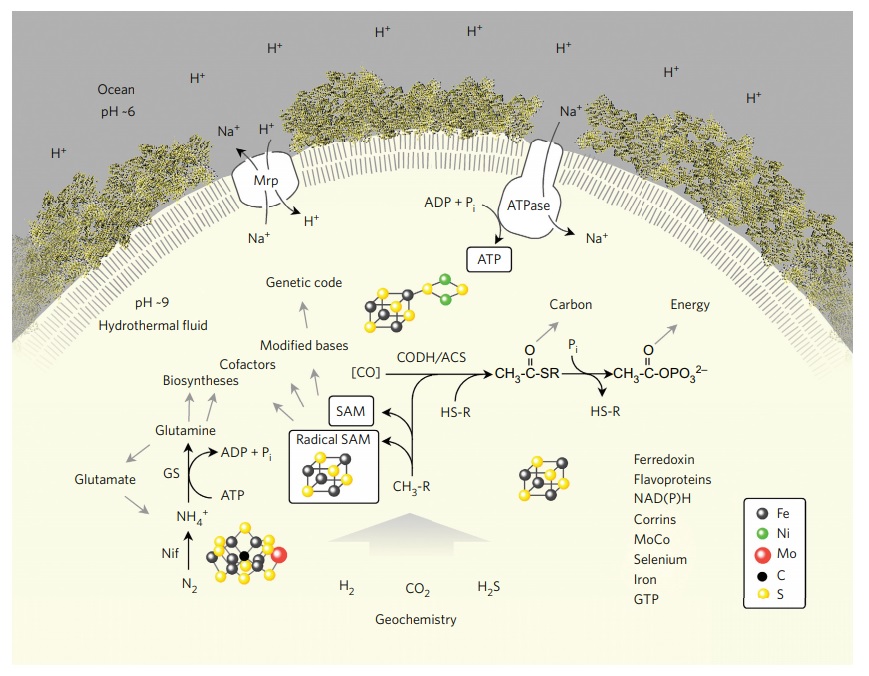
LUCA reconstructed from genome data.
Summary of the main interactions of LUCA with its environment, a vent-like geochemical setting,. Abbreviations: CODH/ACS, carbon monoxide dehydrogenase/acetyl CoA-synthase; Nif, nitrogenase; GS, glutamine synthetase; Mrp, MrP type Na+ /H+ antiporter; CH3-R, methyl groups; HS-R, organic thiols. The components listed on the lower right are present in LUCA. In modern CODH/ACS complexes, CO is generated from CO2 and reduced ferredoxin21. The figure does not make a statement regarding the source of CO in primordial metabolism (uncatalysed via the gas water shift reaction or catalysed via transition metals), symbolized by [CO]. A Na+ /H+ antiporter could transduce a geochemical pH gradient (indicated on the left) inherent in alkaline hydrothermal vents into a more stable Na+ gradient to feed a primordial Na-dependent ATP synthase. LUCA undisputably possessed genes, because it had a genetic code; the question of which genes it possessed has hitherto been more difficult to address. The transition metal catalysts at the nitrogenase active site and the CODH/ACS active site as well as a 4Fe–4S cluster as in ferredoxin are indicated.
Tracing proteins to LUCA by removing transdomain LGTs. Using the standard Markov cluster algorithm (MCL) at a 25% global identity threshold, we sorted all protein coding genes in 1,847 bacterial and 134 archaeal genomes into 286,514 protein families, or clusters, 11,093 of which contained homologues from bacteria and archaea. After alignment and maximum likelihood (ML) tree construction, only 355 clusters preserve domain monophyly while also having homologues in ≥2 archaeal lineages and ≥2 bacterial lineages. Encouragingly, 83% (294/355) of LUCA’s genes have some functional annotation, with only a minority belonging to translation. These 355 proteins were probably present in LUCA and thus provide a glimpse of LUCA’s genome. However, there are also quality benchmarks against which to check the list. For example, LUCA’s genes encode 19 proteins involved in ribosome biogenesis and eight aminoacyl tRNA synthetases, which are also essential for the genetic code to work. Thus, our phylogenetic criteria do not miss the informational core, which itself can be affected by LGTs, such that only subsets of even universally present genes will also meet the domain monophyly criterion. As another benchmark, our phylogenetic criteria return a highly non-random sample of genes.
LUCA’s microbial ecology reconstructed from genomes. Reconstructed from genomic data, LUCA emerges as an anaerobic autotroph that used a Wood–Ljungdahl (WL) pathway and existed in a hydrothermal setting, but that was only half-alive ( sic ) and was dependent upon geochemistry. LUCA’s genes harbor traces of carbon, energy, and nitrogen metabolism. Cells conserve energy via chemiosmotic coupling with rotor–stator-type ATP synthases or via substrate-level phosphorylation (SLP). LUCA’s genes encompass components of two enzymes of energy metabolism: phosphotransacetylase (PTA) and an ATP synthase subunit. PTA generates acetyl phosphate from acetyl-CoA, conserving the energy in the thioester bond as the energy-rich anhydride bond of acetylphosphate, which can phosphorylate ADP or other substrates. The PTA reaction plays a central role in autotrophic theories of microbial origins that focus on thioester-dependent SLP as the ancestral state of microbial energy metabolism. The presence of a rotor-stator ATP synthase subunit points to LUCA’s ability to harness ion gradients for energy metabolism, yet the rotor–stator ATP synthase has undergone transdomain LGT, excluding many of its subunits from LUCA’s set. Crucially, components of electron-transfer-dependent ion-pumping are altogether lacking among LUCA’s genes. LUCA’s ATPase was possibly able to harness geochemically derived ion gradients via H+ /Na+ antiporters, which are present among the membrane proteins. The presence of reverse gyrase, an enzyme specific for hyperthermophiles, indicates a thermophilic lifestyle for LUCA. Enzymes of chemoorganoheterotrophy are lacking, but enzymes for chemolithoautotrophy are present. Among the six known pathways of CO2 fixation, only enzymes of the WL pathway are present in LUCA. LUCA’s WL enzymes are replete with FeS and FeNiS centres, indicating transition-metal requirements and also requiring organic cofactors: flavin, F420, methanofuran, two pterins (the molybdenum cofactor MoCo and tetrahydromethanopterin) and corrins . Microbes that use the WL pathway obtain their electrons from hydrogen, hydrogenases also being present among LUCA’s genes. LUCA accessed nitrogen via nitrogenase and via glutamine synthetase. The WL pathway, nitrogenase and hydrogenases are also very oxygen-sensitive. LUCA was an anaerobic autotroph that could live from the gases H2, CO2 and N2 Several cofactor biosynthesis pathways trace to LUCA, including those for pterins, MoCo, cobalamin, siroheme, thiamine pyrophosphate, coenzyme M and F420. Many of these enzymes are S-adenosyl methionine (SAM)-dependent. A number of LUCA’s SAM-dependent enzymes are radical SAM enzymes, an ancient class of oxygen-sensitive proteins harboring FeS centers that initiate radical-dependent methylations and a wide spectrum of radical reaction mechanisms. Radical SAM reactions point to a prevalence of one-electron reactions in LUCA’s central metabolism, as does an abundance of flavoproteins, in addition to a prominent role for methyl groups. FeS clusters, long viewed as relics of ancient metabolism, are the second most common cofactor/prosthetic group in LUCA’s proteins behind ATP. The abundance of transition metals and FeS as well as FeNiS clusters in LUCA’s enzymes indicates that it inhabited an environment rich in these metals. These features of LUCA’s environment, in addition to thermophily and H2, clearly point to a hydrothermal setting. Selenoproteins, required in glutathione and thioredoxin synthesis and for some of LUCA’s RNA modifications, are present, as is selenophosphate synthase. Like FeS centers, selenium in amino acids and nucleosides is thought to be an ancient trait. Sulfur was involved in ancient metabolism and LUCA was capable of S utilization, as indicated by siroheme, which is specific to redox reactions involving environmental S. Enzymes for sugar metabolism mainly encompass glycosylases, hydrolases and nonoxidative sugar metabolism, possibly reflecting primitive cell wall synthesis.
LUCA’s genes point to acetogenic and methanogenic roots. When LUCA existed, biological hydrogen H2 production did not exist, because primordial organics delivered from space are non-fermentable substrates. For LUCA, that leaves only geological sources of H2. The main geological source of H2, both today and on the early Earth, is serpentinization, a process in which Fe2+ in the crust reduces water circulating through hydrothermal systems to produce H2 at high activities in hydrothermal effluent of up to 26 mmol kg–1 . Yet, as well as H2, methane and other reduced C1 compounds are synthesized abiotically in hydrothermal systems today.
Hydrothermal vents, methyl groups, and nucleoside modifications. LUCA’s genes for RNA nucleoside modification indicate that it performed chemical modification of nucleosides in both tRNA and rRNA34. Four of LUCA’s nucleoside modifications are methylations requiring SAM. In the modern code, several base modifications are even strictly required for codon-anticodon interactions at the wobble position. Consistent with the recurrent role of methyl groups in LUCA’s biology, by far the most common tRNA and rRNA nucleoside modifications that are conserved across the archaeal bacterial divide are methylations, although thiomethylations and incorporation of sulfur and selenium are observed. That LUCA’s genetic code involved modified bases in tRNA– mRNA–rRNA interactions attribute antiquity and functional significance to methylated bases in the evolution of the ribosome and the genetic code. It also forges links between the genetic code, primitive carbon and energy metabolism and hydrothermal environments. How so? At modern hydrothermal vents, reduced C1 intermediates are formed during the abiotic synthesis of methane. The intermediates can accumulate in some modern hydrothermal systems and the underlying reactions can be simulated in the laboratory. These reactions occur because under the reducing conditions of hydrothermal vents, the equilibrium in the reaction of H2 with CO2 lies on the side of reduced carbon compounds. That is the reason why methanogens and clostridial acetogens, both of which figure prominently in autotrophic theories for life’s origin, can grow by harnessing energy from the exergonic reactions of hydrogenotrophic methane and acetate synthesis. The genes in LUCA’s list and the basal lineages among the 355 reciprocally rooted trees indicate that LUCA lived in an environment where the geochemical synthesis of methane from H2 and CO2 was taking place, hence where chemically accessible reduced C1 intermediates existed. Where LUCA arose, the genetic code arose. Either the chemical modifications of RNA nucleosides were absent in LUCA and were introduced later in evolution by some kind of adaptation, as some have suggested, or they are ancient. Conservation of nucleoside modifications across the archaeal bacterial divide indicates the latter. The enzymes that introduce nucleoside methylations are typically SAM enzymes, including members of the radical SAM family, which harbour an FeS cluster that initiates a radical in the reaction mechanism and which are currently thought to be among the most ancient enzymes in metabolism. Both the presence in LUCA’s genome of several SAM enzymes involved in nucleoside modifications and the presence of the nucleosides themselves, sometimes even at conserved positions in tRNA, indicate that these nucleoside methylations were present in LUCA’s code, reflecting the code’s ancestral state. In methanogens and acetogenic clostridia, which the trees identified as the closest relatives of LUCA, methyl groups are central to growth, comprising the very core of carbon and energy metabolism. the methyl group generated by the WL pathway in the energy metabolism of methanogens is transferred from a nitrogen atom in tetrahydromethanopterin to a Co(I) atom in a corrin cofactor of the methyltransferase (MtrA-H) complex, possibly bound by MtrE44, then subsequently transferred to the thiol sulfur atom of coenzyme M before being transferred to hydride at the methyl–CoM reductase reaction. Energy conservation via Na+ pumping occurs during the N-to-Co(I)-to-S transfer sequences of the MtrA-H complex. This is an unusual coupling reaction in that electrons are not transferred; a methyl group is instead transferred, from a N atom to a S atom. The methyl transfer chain of acetogens is a bit longer. It starts with a nitrogen-bound methyl moiety in tetrahydrofolate, which is transferred by AscE to a Co(I) atom in a corrin cofactor of the corrinoid FeS protein and onto a Ni atom in the FeNiS cluster of acetyl-CoA synthase in an unusual metal-tometal methyltransferase reaction. Carbonyl insertion generates a Ni-bound acetyl group that is removed via thiolysis to generate the thioester, which can either be used for carbon assimilation or for energy conservation as acyl phosphate and ATP. In methanogens, carbon metabolism follows the same path to the thioester. These methyl transfer reactions suggest that the environment where primordial carbon and energy metabolism arose was rich in methyl groups, S and transition metals. The conserved SAMdependent methylations and S substitutions in modified nucleosides that allow tRNA anticodons to decode mRNA into protein carry the same chemical imprint, uncovering a hitherto underappreciated antiquity and significance of methyl groups at the core of biological chemistry. Methyl groups provide previously unrecognized links between carbon and energy metabolism in anaerobic autotrophs, tRNA–mRNA–rRNA interactions in the genetic code and spontaneous chemistry at hydrothermal vents
Spelling out caveats and allowing for some LGT. No approach to the study of early evolution is consummate; there are always caveats. Using our strict phylogenetic criterion, 355 protein families that are present in at least two higher taxa per domain and that preserve interdomain monophyly were identified. Universally distributed genes can be subject to transdomain LGT, yielding false negatives and underestimates of LUCA’s gene content, while multiple LGT events might mimic vertical inheritance for some clusters, yielding false positives, or overestimates of LUCA’s gene content. As an example of the latter, O2-dependent enzymes should generally be absent from the list, because in LUCA’s day, O2 did not exist in physiologically relevant amounts. LUCA’s list does, however, contain five enzymes that use O2 as a substrate and three that detoxify O2, functions that cannot be germane to LUCA, hence resulting from multiple transfers that phylogenetically emulate vertical intradomain inheritance. Given the massive influence that O2 had on the origin and spread of new genes during evolution, finding O2-dependent reactions at a frequency of 2.3% (8/355 proteins) suggests that the list of 355 genes harbor comparatively few multiple transfer cases. At the same time, ecological specialization to oxic niches will induce massive loss of anaerobe-specific genes in many lineages, leading to an underestimation of LUCA’s gene content. In addition, LUCA’s gene list reveals only nine nucleotide biosynthesis and five amino acid biosynthesis proteins. The paucity of enzymes for essential amino acid, nucleoside and cofactor biosyntheses is most easily attributed to three factors:
(1) the missing genes in question have been subject to interdomain LGT;
(2) the genes are not well conserved at the sequence level, such that the bacterial and archaeal homologues do not fall into the same cluster;
(3) LUCA had not yet evolved the genes in question prior to the bacterial–archaeal split, the pathway products for LUCA being provided by primordial geochemistry instead.
Low sequence conservation for proteins that were present in LUCA can also yield underestimates of LUCA’s gene content. Because our criteria for presence in LUCA (domain monophyly) involve trees, the sequences need to be sufficiently well conserved to permit multiple sequence alignments and ML phylogenies, so a clustering threshold of 25% global identity was used. Yet genes that were present in LUCA that were not subject to transdomain LGT, but are not well conserved, can still fall into separate domain specific clusters. Relative to archaea, bacteria are overrepresented in the 355 families by a ratio of 134:1,847 in terms of genome sequences. Despite our phylogenetic criteria, some LGTs might be among them. Enzymes for lipid metabolism in LUCA are scarce. The presence of a few enzymes involved in acyl-CoA metabolism might reflect multiple LGTs, as several archaea have acquired bacterial genes for fatty acid and aliphatic degradations. Finally, one might ask what happens if we allow for a little bit of transdomain LGT? The minimum amount of transdomain LGT for which to allow would be reflected by a tree that fulfils our criteria of being present in two members each in two archaeal and two bacterial phyla (see Methods), but in addition, one bacterial sequence is misplaced within the archaea (or vice versa). If we allow for such cases representing one single transdomain LGT, then 124 new trees would be included, expanding our list to 479 members. If we allow for the next increment of LGT, namely that not one sequence but sequences from one archaeal phylum are misplaced within the bacteria (or vice versa), then 97 additional trees would be included, bringing the list to 576 proteins.
Discussion Our findings clearly support the views that FeS and transition metals are relics of ancient metabolism, that life arose at hydrothermal vents, that spontaneous chemistry in the Earth’s crust driven by rock–water interactions at disequilibrium thermodynamically underpinned life’s origin and that the founding lineages of the archaea and bacteria were H2-dependent autotrophs that used CO2 as their terminal acceptor in energy metabolism. Spontaneous reactions involving C1 compounds and intermediate methyl groups in modern submarine hydrothermal systems link observable geochemical processes with the earliest forms of carbon and energy metabolism in bacteria and archaea. In the same way that biochemists have long viewed FeS clusters as relics of ancient catalysis, methyl groups appear here as relics of primordial carbon and energy metabolism. Although the paucity in LUCA of genes for amino acid and nucleoside biosyntheses could, in principle, be attributable to post-LUCA LGT, we note that there is no viable alternative to the view that LUCA, regardless of how envisaged, ultimately arose from components that were synthesized abiotically via spontaneous, exergonic syntheses somewhere during the history of early Earth.
In this paper
The physiology and habitat of the last universal common ancestor
https://sci-hub.tw/https://www.nature.com/articles/nmicrobiol2016116
published by Nature magazine in 2016, they propose a Last universal common ancestor as our ur-progenitor, as the first life form from which all species evolved. They list 355 protein families which i list in the link.
https://reasonandscience.catsboard.com/t2980-the-physiology-and-habitat-of-the-last-universal-common-ancestor#7686
That gives un an idea of mount unsurmountable that supposedly unguided random events had to overcome, to get a first living self replicating cell. It is insane to believe, that such kind of complexity would emerge unguided, by a freaky accident deep in the past.
Consider that all this complexity had to emerge all at once to be functional. A step-wise chemical evolutionary process is not feasible.
https://reasonandscience.catsboard.com/t2980-the-physiology-and-habitat-of-the-last-universal-common-ancestor
Evolution has been tested and it fails. 60,000 generations of bacteria, and all they got, are bacteria. No hint of a transition zone to a new organismal limb or improvement of complexity. Fail.
25 JULY 2016
The concept of a last universal common ancestor of all cells (LUCA, or the progenote) is central to the study of early evolution and life’s origin, yet information about how and where LUCA lived is lacking. Because these proteins are not universally distributed, they can shed light on LUCA’s physiology. Their functions, properties and prosthetic groups depict LUCA as
anaerobic, CO2-fixing, H2-dependent with a Wood–Ljungdahl pathway, N2-fixing and thermophilic.
The list with 355 protein annotations is provided by supplementary table 2 near the end of the page:
https://www.nature.com/articles/nmicrobiol2016116
Ribosome biogenesis
Translation
RNA modification
DNA binding
Nucleic acid handling
Energy metabolism
Carbon assimilation
Nitrogen assimilation
Cofactor biosynthesis
Nucleotide metabolism
Amino acid metabolism
Redox chemistry
Protein modification
Lipid metabolism
Sugar-related
Cellular
Cell wall-related
Transport
Others
Unknown & uncharacterized & predicted
Oxygen
The cell is irreducibly complex
https://reasonandscience.catsboard.com/t1299-abiogenesis-the-cell-is-irreducibly-complex
Biological Cells are irreducibly complex:
1. Cell subunits and compartments form a complex system that is useful only in the completion of a much larger system that is able to keep the basic functions of life. A minimal Cell, in order to permit life, according to an article in Science magazine, from 2016, requires a minimal genome of about 473gene products, and at least 438 proteins, each fully set up and functional for specific tasks. A discrete minimal size of each individual protein complex formed by multiple subunits and cofactors is required in order to be functional. And it only operates when interconnected and in a joint venture similar to a robot in a production line, and precise energy supply.
2. Cells must be created and be functional, all at once. As Graham Cairns-Smith noted, this system has to be fixed in its essentials through the critical interdependence of subsystems. Irreducibly complex and interdepend systems cannot evolve but depend on intelligence with foreknowledge on how to build discrete parts with distant goals.
3. Therefore, intelligent design is the best explanation of the origin of living, self-replicating cells.
Chemistry and the Missing Era of Evolution: A. Graham Cairns-Smith
We can see that at the time of the common ancestor, this system must already have been fixed in its essentials, probably through a critical interdependence of subsystems. (Roughly speaking in a domain in which everything has come to depend on everything else nothing can be easily changed, and our central biochemistry is very much like that.
https://sci-hub.tw/https://www.ncbi.nlm.nih.gov/pubmed/18260066
The role of natural selection in the origin of life
Unlike living systems that are products of and participants in evolution, these prebiotic chemical structures were not products of evolution. Not being yet intricately organized, they could have emerged as a result of ordinary physical and chemical processes.
https://www.ncbi.nlm.nih.gov/pubmed/20407927
chemist Wilhelm Huck, professor at Radboud University Nijmegen
A working cell is more than the sum of its parts. "A functioning cell must be entirely correct at once, in all its complexity
http://www.ru.nl/english/@893712/protocells-formed/
https://www.sciencedaily.com/releases/2013/07/130702100115.htm
How Structure Arose in the Primordial Soup
About 4 billion years ago, molecules began to make copies of themselves, an event that marked the beginning of life on Earth. A few hundred million years later, primitive organisms began to split into the different branches that make up the tree of life. In between those two seminal events, some of the greatest innovations in existence emerged: the cell, the genetic code and an energy system to fuel it all. ALL THREE of these are ESSENTIAL to life as we know it, yet scientists know disappointingly little about how any of these remarkable biological innovations came about.
https://www.scientificamerican.com/article/how-structure-arose-in-the-primordial-soup/
We investigated all clusters and phylogenetic trees for 6.1 million protein-coding genes from sequenced prokaryotic genomes in order to reconstruct the microbial ecology of LUCA. Among 286,514 protein clusters, we identified 355 protein families (∼0.1%) that trace to LUCA by phylogenetic criteria.
LUCA’s biochemistry was replete with FeS clusters and radical reaction mechanisms. Its cofactors reveal dependence upon transition metals, flavins, S-adenosyl methionine, coenzyme A, ferredoxin, molybdopterin, corrins and selenium. Its genetic code required nucleoside modifications and S-adenosyl methionine-dependent methylations. The 355 phylogenies identify clostridia and methanogens, whose modern lifestyles resemble that of LUCA, as basal among their respective domains. LUCA inhabited a geochemically active environment rich in H2, CO2 and iron. The data support the theory of an autotrophic origin of life involving the Wood–Ljungdahl pathway in a hydrothermal setting.
The last universal common ancestor (LUCA) is an inferred evolutionary intermediate that links the abiotic phase of Earth’s history with the first traces of microbial life in rocks that are 3.8–3.5 billion years of age . Although LUCA was long considered the common ancestor of bacteria, archaea and eukaryotes newer two-domain trees of life have eukaryotes arising from prokaryotes making LUCA the common ancestor of bacteria and archaea. Previous genomic investigations of LUCA’s gene content have focused on genes that are universally present across genomes, LUCA had 30–100 proteins for ribosomes and translation. To identify genes that can illuminate the biology of LUCA, we took a phylogenetic approach. By focusing on the phylogeny rather than universal gene presence, we can identify genes involved in LUCA’s physiology—the ways that cells access carbon, energy and nutrients from the environment for growth.

LUCA reconstructed from genome data.
Summary of the main interactions of LUCA with its environment, a vent-like geochemical setting,. Abbreviations: CODH/ACS, carbon monoxide dehydrogenase/acetyl CoA-synthase; Nif, nitrogenase; GS, glutamine synthetase; Mrp, MrP type Na+ /H+ antiporter; CH3-R, methyl groups; HS-R, organic thiols. The components listed on the lower right are present in LUCA. In modern CODH/ACS complexes, CO is generated from CO2 and reduced ferredoxin21. The figure does not make a statement regarding the source of CO in primordial metabolism (uncatalysed via the gas water shift reaction or catalysed via transition metals), symbolized by [CO]. A Na+ /H+ antiporter could transduce a geochemical pH gradient (indicated on the left) inherent in alkaline hydrothermal vents into a more stable Na+ gradient to feed a primordial Na-dependent ATP synthase. LUCA undisputably possessed genes, because it had a genetic code; the question of which genes it possessed has hitherto been more difficult to address. The transition metal catalysts at the nitrogenase active site and the CODH/ACS active site as well as a 4Fe–4S cluster as in ferredoxin are indicated.
Tracing proteins to LUCA by removing transdomain LGTs. Using the standard Markov cluster algorithm (MCL) at a 25% global identity threshold, we sorted all protein coding genes in 1,847 bacterial and 134 archaeal genomes into 286,514 protein families, or clusters, 11,093 of which contained homologues from bacteria and archaea. After alignment and maximum likelihood (ML) tree construction, only 355 clusters preserve domain monophyly while also having homologues in ≥2 archaeal lineages and ≥2 bacterial lineages. Encouragingly, 83% (294/355) of LUCA’s genes have some functional annotation, with only a minority belonging to translation. These 355 proteins were probably present in LUCA and thus provide a glimpse of LUCA’s genome. However, there are also quality benchmarks against which to check the list. For example, LUCA’s genes encode 19 proteins involved in ribosome biogenesis and eight aminoacyl tRNA synthetases, which are also essential for the genetic code to work. Thus, our phylogenetic criteria do not miss the informational core, which itself can be affected by LGTs, such that only subsets of even universally present genes will also meet the domain monophyly criterion. As another benchmark, our phylogenetic criteria return a highly non-random sample of genes.
LUCA’s microbial ecology reconstructed from genomes. Reconstructed from genomic data, LUCA emerges as an anaerobic autotroph that used a Wood–Ljungdahl (WL) pathway and existed in a hydrothermal setting, but that was only half-alive ( sic ) and was dependent upon geochemistry. LUCA’s genes harbor traces of carbon, energy, and nitrogen metabolism. Cells conserve energy via chemiosmotic coupling with rotor–stator-type ATP synthases or via substrate-level phosphorylation (SLP). LUCA’s genes encompass components of two enzymes of energy metabolism: phosphotransacetylase (PTA) and an ATP synthase subunit. PTA generates acetyl phosphate from acetyl-CoA, conserving the energy in the thioester bond as the energy-rich anhydride bond of acetylphosphate, which can phosphorylate ADP or other substrates. The PTA reaction plays a central role in autotrophic theories of microbial origins that focus on thioester-dependent SLP as the ancestral state of microbial energy metabolism. The presence of a rotor-stator ATP synthase subunit points to LUCA’s ability to harness ion gradients for energy metabolism, yet the rotor–stator ATP synthase has undergone transdomain LGT, excluding many of its subunits from LUCA’s set. Crucially, components of electron-transfer-dependent ion-pumping are altogether lacking among LUCA’s genes. LUCA’s ATPase was possibly able to harness geochemically derived ion gradients via H+ /Na+ antiporters, which are present among the membrane proteins. The presence of reverse gyrase, an enzyme specific for hyperthermophiles, indicates a thermophilic lifestyle for LUCA. Enzymes of chemoorganoheterotrophy are lacking, but enzymes for chemolithoautotrophy are present. Among the six known pathways of CO2 fixation, only enzymes of the WL pathway are present in LUCA. LUCA’s WL enzymes are replete with FeS and FeNiS centres, indicating transition-metal requirements and also requiring organic cofactors: flavin, F420, methanofuran, two pterins (the molybdenum cofactor MoCo and tetrahydromethanopterin) and corrins . Microbes that use the WL pathway obtain their electrons from hydrogen, hydrogenases also being present among LUCA’s genes. LUCA accessed nitrogen via nitrogenase and via glutamine synthetase. The WL pathway, nitrogenase and hydrogenases are also very oxygen-sensitive. LUCA was an anaerobic autotroph that could live from the gases H2, CO2 and N2 Several cofactor biosynthesis pathways trace to LUCA, including those for pterins, MoCo, cobalamin, siroheme, thiamine pyrophosphate, coenzyme M and F420. Many of these enzymes are S-adenosyl methionine (SAM)-dependent. A number of LUCA’s SAM-dependent enzymes are radical SAM enzymes, an ancient class of oxygen-sensitive proteins harboring FeS centers that initiate radical-dependent methylations and a wide spectrum of radical reaction mechanisms. Radical SAM reactions point to a prevalence of one-electron reactions in LUCA’s central metabolism, as does an abundance of flavoproteins, in addition to a prominent role for methyl groups. FeS clusters, long viewed as relics of ancient metabolism, are the second most common cofactor/prosthetic group in LUCA’s proteins behind ATP. The abundance of transition metals and FeS as well as FeNiS clusters in LUCA’s enzymes indicates that it inhabited an environment rich in these metals. These features of LUCA’s environment, in addition to thermophily and H2, clearly point to a hydrothermal setting. Selenoproteins, required in glutathione and thioredoxin synthesis and for some of LUCA’s RNA modifications, are present, as is selenophosphate synthase. Like FeS centers, selenium in amino acids and nucleosides is thought to be an ancient trait. Sulfur was involved in ancient metabolism and LUCA was capable of S utilization, as indicated by siroheme, which is specific to redox reactions involving environmental S. Enzymes for sugar metabolism mainly encompass glycosylases, hydrolases and nonoxidative sugar metabolism, possibly reflecting primitive cell wall synthesis.
LUCA’s genes point to acetogenic and methanogenic roots. When LUCA existed, biological hydrogen H2 production did not exist, because primordial organics delivered from space are non-fermentable substrates. For LUCA, that leaves only geological sources of H2. The main geological source of H2, both today and on the early Earth, is serpentinization, a process in which Fe2+ in the crust reduces water circulating through hydrothermal systems to produce H2 at high activities in hydrothermal effluent of up to 26 mmol kg–1 . Yet, as well as H2, methane and other reduced C1 compounds are synthesized abiotically in hydrothermal systems today.
Hydrothermal vents, methyl groups, and nucleoside modifications. LUCA’s genes for RNA nucleoside modification indicate that it performed chemical modification of nucleosides in both tRNA and rRNA34. Four of LUCA’s nucleoside modifications are methylations requiring SAM. In the modern code, several base modifications are even strictly required for codon-anticodon interactions at the wobble position. Consistent with the recurrent role of methyl groups in LUCA’s biology, by far the most common tRNA and rRNA nucleoside modifications that are conserved across the archaeal bacterial divide are methylations, although thiomethylations and incorporation of sulfur and selenium are observed. That LUCA’s genetic code involved modified bases in tRNA– mRNA–rRNA interactions attribute antiquity and functional significance to methylated bases in the evolution of the ribosome and the genetic code. It also forges links between the genetic code, primitive carbon and energy metabolism and hydrothermal environments. How so? At modern hydrothermal vents, reduced C1 intermediates are formed during the abiotic synthesis of methane. The intermediates can accumulate in some modern hydrothermal systems and the underlying reactions can be simulated in the laboratory. These reactions occur because under the reducing conditions of hydrothermal vents, the equilibrium in the reaction of H2 with CO2 lies on the side of reduced carbon compounds. That is the reason why methanogens and clostridial acetogens, both of which figure prominently in autotrophic theories for life’s origin, can grow by harnessing energy from the exergonic reactions of hydrogenotrophic methane and acetate synthesis. The genes in LUCA’s list and the basal lineages among the 355 reciprocally rooted trees indicate that LUCA lived in an environment where the geochemical synthesis of methane from H2 and CO2 was taking place, hence where chemically accessible reduced C1 intermediates existed. Where LUCA arose, the genetic code arose. Either the chemical modifications of RNA nucleosides were absent in LUCA and were introduced later in evolution by some kind of adaptation, as some have suggested, or they are ancient. Conservation of nucleoside modifications across the archaeal bacterial divide indicates the latter. The enzymes that introduce nucleoside methylations are typically SAM enzymes, including members of the radical SAM family, which harbour an FeS cluster that initiates a radical in the reaction mechanism and which are currently thought to be among the most ancient enzymes in metabolism. Both the presence in LUCA’s genome of several SAM enzymes involved in nucleoside modifications and the presence of the nucleosides themselves, sometimes even at conserved positions in tRNA, indicate that these nucleoside methylations were present in LUCA’s code, reflecting the code’s ancestral state. In methanogens and acetogenic clostridia, which the trees identified as the closest relatives of LUCA, methyl groups are central to growth, comprising the very core of carbon and energy metabolism. the methyl group generated by the WL pathway in the energy metabolism of methanogens is transferred from a nitrogen atom in tetrahydromethanopterin to a Co(I) atom in a corrin cofactor of the methyltransferase (MtrA-H) complex, possibly bound by MtrE44, then subsequently transferred to the thiol sulfur atom of coenzyme M before being transferred to hydride at the methyl–CoM reductase reaction. Energy conservation via Na+ pumping occurs during the N-to-Co(I)-to-S transfer sequences of the MtrA-H complex. This is an unusual coupling reaction in that electrons are not transferred; a methyl group is instead transferred, from a N atom to a S atom. The methyl transfer chain of acetogens is a bit longer. It starts with a nitrogen-bound methyl moiety in tetrahydrofolate, which is transferred by AscE to a Co(I) atom in a corrin cofactor of the corrinoid FeS protein and onto a Ni atom in the FeNiS cluster of acetyl-CoA synthase in an unusual metal-tometal methyltransferase reaction. Carbonyl insertion generates a Ni-bound acetyl group that is removed via thiolysis to generate the thioester, which can either be used for carbon assimilation or for energy conservation as acyl phosphate and ATP. In methanogens, carbon metabolism follows the same path to the thioester. These methyl transfer reactions suggest that the environment where primordial carbon and energy metabolism arose was rich in methyl groups, S and transition metals. The conserved SAMdependent methylations and S substitutions in modified nucleosides that allow tRNA anticodons to decode mRNA into protein carry the same chemical imprint, uncovering a hitherto underappreciated antiquity and significance of methyl groups at the core of biological chemistry. Methyl groups provide previously unrecognized links between carbon and energy metabolism in anaerobic autotrophs, tRNA–mRNA–rRNA interactions in the genetic code and spontaneous chemistry at hydrothermal vents
Spelling out caveats and allowing for some LGT. No approach to the study of early evolution is consummate; there are always caveats. Using our strict phylogenetic criterion, 355 protein families that are present in at least two higher taxa per domain and that preserve interdomain monophyly were identified. Universally distributed genes can be subject to transdomain LGT, yielding false negatives and underestimates of LUCA’s gene content, while multiple LGT events might mimic vertical inheritance for some clusters, yielding false positives, or overestimates of LUCA’s gene content. As an example of the latter, O2-dependent enzymes should generally be absent from the list, because in LUCA’s day, O2 did not exist in physiologically relevant amounts. LUCA’s list does, however, contain five enzymes that use O2 as a substrate and three that detoxify O2, functions that cannot be germane to LUCA, hence resulting from multiple transfers that phylogenetically emulate vertical intradomain inheritance. Given the massive influence that O2 had on the origin and spread of new genes during evolution, finding O2-dependent reactions at a frequency of 2.3% (8/355 proteins) suggests that the list of 355 genes harbor comparatively few multiple transfer cases. At the same time, ecological specialization to oxic niches will induce massive loss of anaerobe-specific genes in many lineages, leading to an underestimation of LUCA’s gene content. In addition, LUCA’s gene list reveals only nine nucleotide biosynthesis and five amino acid biosynthesis proteins. The paucity of enzymes for essential amino acid, nucleoside and cofactor biosyntheses is most easily attributed to three factors:
(1) the missing genes in question have been subject to interdomain LGT;
(2) the genes are not well conserved at the sequence level, such that the bacterial and archaeal homologues do not fall into the same cluster;
(3) LUCA had not yet evolved the genes in question prior to the bacterial–archaeal split, the pathway products for LUCA being provided by primordial geochemistry instead.
Low sequence conservation for proteins that were present in LUCA can also yield underestimates of LUCA’s gene content. Because our criteria for presence in LUCA (domain monophyly) involve trees, the sequences need to be sufficiently well conserved to permit multiple sequence alignments and ML phylogenies, so a clustering threshold of 25% global identity was used. Yet genes that were present in LUCA that were not subject to transdomain LGT, but are not well conserved, can still fall into separate domain specific clusters. Relative to archaea, bacteria are overrepresented in the 355 families by a ratio of 134:1,847 in terms of genome sequences. Despite our phylogenetic criteria, some LGTs might be among them. Enzymes for lipid metabolism in LUCA are scarce. The presence of a few enzymes involved in acyl-CoA metabolism might reflect multiple LGTs, as several archaea have acquired bacterial genes for fatty acid and aliphatic degradations. Finally, one might ask what happens if we allow for a little bit of transdomain LGT? The minimum amount of transdomain LGT for which to allow would be reflected by a tree that fulfils our criteria of being present in two members each in two archaeal and two bacterial phyla (see Methods), but in addition, one bacterial sequence is misplaced within the archaea (or vice versa). If we allow for such cases representing one single transdomain LGT, then 124 new trees would be included, expanding our list to 479 members. If we allow for the next increment of LGT, namely that not one sequence but sequences from one archaeal phylum are misplaced within the bacteria (or vice versa), then 97 additional trees would be included, bringing the list to 576 proteins.
Discussion Our findings clearly support the views that FeS and transition metals are relics of ancient metabolism, that life arose at hydrothermal vents, that spontaneous chemistry in the Earth’s crust driven by rock–water interactions at disequilibrium thermodynamically underpinned life’s origin and that the founding lineages of the archaea and bacteria were H2-dependent autotrophs that used CO2 as their terminal acceptor in energy metabolism. Spontaneous reactions involving C1 compounds and intermediate methyl groups in modern submarine hydrothermal systems link observable geochemical processes with the earliest forms of carbon and energy metabolism in bacteria and archaea. In the same way that biochemists have long viewed FeS clusters as relics of ancient catalysis, methyl groups appear here as relics of primordial carbon and energy metabolism. Although the paucity in LUCA of genes for amino acid and nucleoside biosyntheses could, in principle, be attributable to post-LUCA LGT, we note that there is no viable alternative to the view that LUCA, regardless of how envisaged, ultimately arose from components that were synthesized abiotically via spontaneous, exergonic syntheses somewhere during the history of early Earth.
In this paper
The physiology and habitat of the last universal common ancestor
https://sci-hub.tw/https://www.nature.com/articles/nmicrobiol2016116
published by Nature magazine in 2016, they propose a Last universal common ancestor as our ur-progenitor, as the first life form from which all species evolved. They list 355 protein families which i list in the link.
https://reasonandscience.catsboard.com/t2980-the-physiology-and-habitat-of-the-last-universal-common-ancestor#7686
That gives un an idea of mount unsurmountable that supposedly unguided random events had to overcome, to get a first living self replicating cell. It is insane to believe, that such kind of complexity would emerge unguided, by a freaky accident deep in the past.
Consider that all this complexity had to emerge all at once to be functional. A step-wise chemical evolutionary process is not feasible.
Last edited by Admin on Sun 19 Jul 2020 - 17:03; edited 8 times in total