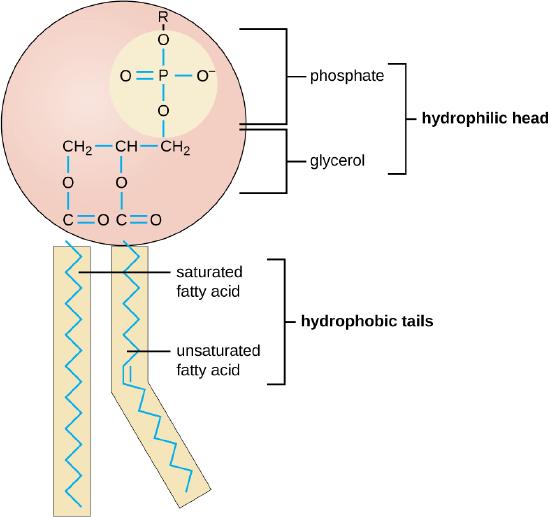
For life to emerge from primordial conditions, a series of critical processes would have had to occur to establish and maintain balanced nucleotide pools:
7.9.1 Spatial Separation Mechanisms for Nucleotide Management
Spatial separation mechanisms would have had to develop to manage nucleotide availability in prebiotic conditions. Several crucial processes would have had to play a vital role in concentrating and protecting these essential molecules. The formation of lipid vesicles would have had to be one such key mechanism, with amphiphilic molecules spontaneously assembling into primitive cell-like structures. These vesicles would have had to provide enclosed environments where nucleotides could accumulate, shielded from the diluting effects of the broader aqueous surroundings. Another important spatial separation strategy would have had to involve the use of mineral surfaces as scaffolds. Clays, zeolites, and other minerals with high surface areas and charged surfaces would have had to adsorb nucleotides, concentrating them and potentially catalyzing their formation or polymerization. This mineral-based approach would have been particularly relevant in geologically active areas, where fresh mineral surfaces were constantly exposed. Isolated microenvironments in porous rock formations would have had to provide another means of spatial separation. These natural compartments, formed by the intricate network of cavities and channels in certain rock types, would have acted as primitive reaction vessels. Within these spaces, nucleotides would have had to accumulate to higher concentrations than in the open environment, facilitating more complex chemical reactions. These separated spaces—whether lipid vesicles, mineral surfaces, or rock pores—would have had to be crucial for allowing localized nucleotide concentration. This concentration effect would have had to dramatically increase the likelihood of nucleotides interacting with each other and with other prebiotic molecules, potentially leading to the formation of longer RNA or DNA sequences. Moreover, these compartmentalized environments would have had to play a vital role in protecting nucleotides from dilution in the broader environment. Given the vast volumes of the primordial oceans, maintaining sufficient concentrations of these complex molecules would have been a significant challenge. The spatial separation mechanisms would have had to provide a solution to this dilution problem, creating pockets of high nucleotide concentration that could persist over time. These mechanisms would not have operated in isolation but would have had to interact and complement each other. For instance, lipid vesicles could have formed within the pores of rocks, or mineral particles could have been incorporated into the membranes of vesicles, creating even more complex and potentially more effective spatial separation systems. The development of these spatial separation mechanisms would have been a critical step toward the origin of life, providing the necessary conditions for nucleotide concentration and protection, and setting the stage for the emergence of more complex biological systems.
A 2023 study by Himbert et al. investigates the role of mineral surfaces in the formation of prebiotic RNA polymers. It is claimed that mineral surfaces like clays and metal oxides were crucial in concentrating nucleotides, offering a solution to the dilution problem in the primordial oceans. These surfaces facilitated the organization and potential polymerization of nucleotides by providing adsorption sites that helped localize molecules, increasing their interactions. However, the study suggests that while mineral surfaces organized nucleotides into structured arrangements, they did not catalyze the formation of pre-polymers directly. Instead, it is hypothesized that wet-dry cycles, where water levels fluctuated, likely played a key role in promoting the formation of hydrogen bonds between nucleotides, leading to early polymerization. This research provides a plausible mechanism for spatial separation and nucleotide management in prebiotic conditions, focusing on how mineral surfaces and compartmentalization could have concentrated essential molecules for early life. This supports the hypothesis that mineral surfaces, along with lipid vesicles and other microenvironments, created the conditions necessary for nucleotide accumulation and protection, setting the stage for more complex biological systems. 7
Problems Identified:
1. While mineral surfaces promote nucleotide organization, they do not directly catalyze polymer formation.
2. The mechanism of polymerization still relies on external factors like wet-dry cycles, leaving uncertainties in fully explaining prebiotic RNA formation.
3. No definitive model yet shows how these processes could lead to the spontaneous emergence of life from non-living chemistry.
Unresolved Challenges and Conceptual Questions
1. Vesicle Formation and Stability
The spontaneous formation of stable lipid vesicles in prebiotic conditions remains a significant challenge. Current hypotheses struggle to explain how amphiphilic molecules could self-assemble into vesicles capable of selective permeability and long-term stability without guided processes.
Conceptual problem: Spontaneous Membrane Organization
- No known physical principle necessitates the formation of stable, selectively permeable membranes
- Difficulty in explaining the emergence of complex lipid compositions required for membrane functionality
2. Mineral Surface Catalysis and Adsorption
While mineral surfaces are proposed as catalysts and scaffolds for nucleotide concentration, their efficiency and specificity raise substantial questions. The non-specific nature of adsorption and limited catalytic activity observed in experiments challenge the idea of minerals as effective concentrators of nucleotides.
Conceptual problem: Selective Adsorption and Catalysis
- Lack of mechanisms for achieving specific adsorption of nucleotides over other organic molecules
- No clear explanation for how mineral surfaces could catalyze complex reactions with precision
3. Microenvironment Formation in Porous Rocks
The hypothesis that porous rock formations could create isolated microenvironments for nucleotide concentration faces several challenges. There is no clear mechanism for selective accumulation of nucleotides while excluding other substances.
Conceptual problem: Selective Accumulation
- Absence of known physical principles that would allow for preferential concentration of nucleotides in rock pores
- Difficulty in explaining protection from degradation in potentially harsh microenvironments
4. Overcoming Dilution in Primordial Oceans
Maintaining high local concentrations of nucleotides in vast aqueous environments presents a formidable challenge. No convincing mechanism has been proposed for overcoming the constant dilution effect in large bodies of water.
Conceptual problem: Concentration Against Entropy
- Lack of plausible mechanisms for concentrating molecules against thermodynamic gradients
- No clear explanation for how localized high concentrations could be maintained without active processes
5. Interplay Between Different Separation Mechanisms
The potential interaction and complementarity between various spatial separation mechanisms (vesicles, mineral surfaces, rock pores) remain unexplained. There's no clear pathway for how these different environments could have emerged and functioned together coherently.
Conceptual problem: Coordinated Emergence
- Absence of known principles that would necessitate the co-emergence of complementary separation mechanisms
- Difficulty in explaining how different mechanisms could integrate functionally without guidance
6. Protection from Environmental Degradation
The harsh conditions of early Earth pose a significant threat to nucleotide stability. Current hypotheses struggle to explain how spatial separation mechanisms could effectively protect these molecules from degradation.
Conceptual problem: Molecular Preservation
- No clear mechanism for shielding nucleotides from various degradation pathways in prebiotic environments
- Difficulty in explaining how protective environments could emerge and persist without sophisticated maintenance systems
7. Energy Requirements for Concentration
Concentrating nucleotides against concentration gradients requires energy input. In the absence of metabolic processes, it's unclear how this energy could have been consistently provided and harnessed.
Conceptual problem: Energy Coupling
- Lack of plausible mechanisms for coupling environmental energy sources to concentration processes
- Difficulty in explaining sustained energy input required for ongoing nucleotide management
8. Selectivity in Molecular Transport
Effective nucleotide management would require selective transport mechanisms to concentrate specific molecules while excluding others. The emergence of such selectivity without pre-existing biological machinery is problematic.
Conceptual problem: Molecular Recognition
- No known chemical principle that would lead to the spontaneous development of selective transport
- Difficulty in explaining the origin of specific molecular recognition capabilities
9. Temporal Coordination of Separation Processes
The timing and synchronization of various spatial separation mechanisms present another challenge. How could these processes have emerged and operated in a coordinated manner without centralized control?
Conceptual problem: Spontaneous Synchronization
- Absence of known principles that would lead to the temporal coordination of multiple, distinct processes
- Difficulty in explaining how a coherent system of separation mechanisms could emerge from chaotic prebiotic conditions
10. Transition to Self-Replicating Systems
Even if spatial separation mechanisms could concentrate nucleotides, the transition to self-replicating systems remains unexplained. No clear pathway has been demonstrated for how concentrated nucleotides could spontaneously organize into functional, self-replicating entities.
Conceptual problem: Emergence of Self-Replication
- Lack of known chemical principles that necessitate the formation of self-replicating systems from concentrated monomers
- Difficulty in explaining the origin of the information content required for self-replication
These unresolved challenges and conceptual problems highlight the significant gaps in our understanding of how spatial separation mechanisms for nucleotide management could have emerged through purely naturalistic processes. The lack of plausible explanations for these fundamental issues necessitates a critical reevaluation of current hypotheses regarding the origin of life and the capabilities of unguided physical and chemical processes.
7.9.2 Formation of Chemical Gradients for Nucleotide Separation
Chemical gradients would have had to play a crucial role in the prebiotic separation and concentration of nucleotides, creating distinct environments that favored nucleotide retention and synthesis. pH gradients across primitive membranes would have needed to lead to differential ion concentrations, driving the selective accumulation of nucleotides. These pH differences would have had to arise from the inherent properties of early membrane-forming molecules or the action of primitive proton pumps. Charge-based separation mechanisms, leveraging the negative charge of phosphate groups in nucleotides, would have required positively charged surfaces or molecules that selectively interacted with nucleotides, allowing for their concentration and retention. Temperature gradients in geothermal environments, particularly near hydrothermal vents or in shallow pools subject to solar heating, would have had to create convection currents and zones of varying reactivity. These thermal variations would have influenced reaction rates and the stability of different molecular species, favoring nucleotide formation in specific thermal niches. Concentration gradients of metal ions and other catalytic species would have had to develop, creating regions where nucleotide synthesis was more favorable due to the differential solubility and reactivity of various mineral components in the primordial environment. Redox gradients, especially at interfaces between reducing and oxidizing environments, would have been necessary to provide the electron flow required for prebiotic reactions, influencing the oxidation state of key molecular species involved in nucleotide synthesis. Osmotic gradients across primitive membranes would have contributed to the concentration of nucleotides and their precursors within protocellular structures by driving the selective uptake of certain molecules while excluding others. Interfacial gradients at the boundaries between different phases (e.g., liquid-solid, liquid-gas) would have created unique chemical environments conducive to nucleotide formation and retention, offering surfaces for adsorption and catalysis, as well as regions of altered molecular orientation and reactivity. The formation and maintenance of these various chemical gradients would have had to be a dynamic process, driven by environmental energy inputs such as solar radiation, geothermal heat, or chemical disequilibria. The interplay between these different types of gradients would have created a complex, heterogeneous prebiotic environment, providing numerous microenvironments where different stages of nucleotide synthesis and retention could occur optimally. The establishment of these chemical gradients would have been essential for the spatial and functional organization of prebiotic chemistry, paving the way for the emergence of more complex, self-sustaining chemical systems that eventually led to the origin of life.
Unresolved Issues and Conceptual Problems
1. Spontaneous Formation of pH Gradients:
The emergence of pH gradients across primitive membranes presents significant challenges. No known chemical principles necessitate the spontaneous formation of stable pH gradients. Explaining how such gradients could form and maintain themselves without sophisticated biological machinery, such as proton pumps requiring complex proteins, is difficult.
2. Charge-Based Separation Mechanisms:
The selective interaction between positively charged surfaces and the phosphate groups of nucleotides raises questions about specificity and efficiency in prebiotic conditions. There is a lack of mechanisms for achieving specific interactions with nucleotides over other charged molecules. It is unclear how charge-based separation could occur efficiently without interfering side reactions or without the presence of selective molecular recognition systems.
3. Temperature Gradient Formation and Stability:
While temperature gradients can occur naturally, their ability to create and maintain specific zones conducive to nucleotide formation is questionable. It is difficult to explain how stable thermal niches could persist in dynamic prebiotic environments. There is a lack of evidence for how temperature gradients could selectively favor complex nucleotide chemistry over other competing reactions.
4. Metal Ion and Catalytic Species Gradients:
The formation of concentration gradients of metal ions and other catalytic species faces challenges in explaining their stability and specificity. There is no clear mechanism for maintaining localized high concentrations of specific ions in open systems. It is difficult to explain how these gradients could persist without constant external input or how they could selectively promote nucleotide synthesis.
5. Redox Gradient Emergence:
The formation of redox gradients, particularly at oxidizing-reducing interfaces, presents challenges in terms of stability and energy coupling. There is a lack of plausible mechanisms for maintaining stable redox gradients without biological systems. It is difficult to explain how primitive chemical systems could harness electron flow for complex reactions necessary for nucleotide synthesis.
6. Osmotic Gradient Formation Across Primitive Membranes:
The emergence of osmotic gradients capable of concentrating nucleotides within protocellular structures faces significant hurdles. There is no known principle that would lead to the spontaneous development of selectively permeable membranes. It is difficult to explain how osmotic gradients could be maintained without active transport mechanisms or membrane proteins.
7. Interfacial Gradient Complexity:
The formation of complex interfacial gradients conducive to nucleotide synthesis and retention remains unexplained. There is a lack of mechanisms for spontaneously generating and maintaining complex multi-phase interfaces. It is unclear how these interfaces could provide consistent catalytic environments without specific organizational structures.
8. Energy Input for Gradient Maintenance:
The continuous energy input required to maintain various chemical gradients presents a significant challenge in prebiotic scenarios. It is difficult to explain how environmental energy sources could be consistently harnessed without sophisticated molecular machinery. There is a lack of plausible mechanisms for coupling diverse energy inputs to specific gradient-maintaining processes.
9. Gradient Interplay and Microenvironment Formation:
The coordinated interplay between different types of gradients to create suitable microenvironments for nucleotide chemistry remains unexplained. There is no known principle that would necessitate the coordinated emergence of multiple, complementary gradients. It is difficult to explain how diverse gradients could self-organize into functional microenvironments without guided processes.
10. Transition to Self-Sustaining Systems:
Even if chemical gradients could form, the transition to self-sustaining, replicating systems remains a profound mystery. There is a lack of known chemical principles that would lead to the spontaneous development of self-replicating systems from gradient-driven chemistry. It is difficult to explain the origin of the information content and catalytic capability required for self-sustenance.
These unresolved challenges highlight significant gaps in our understanding of how chemical gradients for nucleotide separation could have emerged through purely naturalistic processes. The complexity and precision required for these gradient-based systems suggest that alternative explanations may need to be considered to address these persistent and profound scientific questions.
7.9.3 Development of Selective Permeability in Early Membranes or Barriers
The development of selective permeability in early membranes would have required mechanisms to allow certain molecules to pass while retaining nucleotides and other essential components. Size-based exclusion would have played a key role, with small molecules like water and gases diffusing through while larger, complex molecules would be blocked. Charge-based interactions would have contributed as well, with the membrane's chemical composition determining its affinity for charged or polar molecules, allowing selective ion movement. Additionally, primitive transport systems would have emerged, potentially involving pore-forming structures or simple carrier molecules, facilitating the controlled exchange of nutrients, ions, and metabolites. These membranes would have needed to maintain a balance between permeability and protection, ensuring the internal environment could sustain essential chemical processes while preventing the uncontrolled loss of crucial components. This selective permeability would have been fundamental to the compartmentalization of early cells, allowing metabolic pathways to function effectively and setting the stage for more sophisticated biological membranes in evolving life forms. The development of these selectively permeable barriers would have necessitated the emergence of specific lipid compositions. Amphiphilic molecules capable of self-assembly into bilayer structures would have had to form spontaneously in the prebiotic environment. These early membranes would have required a degree of fluidity to allow for the insertion and movement of primitive transport molecules, while still maintaining structural integrity. Membrane asymmetry would have had to develop, with different lipid compositions on the inner and outer leaflets of the membrane. This asymmetry would have been crucial for establishing directional transport and creating distinct internal and external environments. The incorporation of primitive proteins or peptides into these early membranes would have been necessary for enhancing selective permeability. These proteinaceous components would have formed rudimentary channels or pores, allowing for more specific and controlled molecular transport. Mechanisms for membrane repair and growth would have had to evolve concurrently. The ability to incorporate new lipid molecules and expand the membrane surface area would have been essential for the growth and division of early protocells. The development of proton gradients across these early membranes would have been a critical step. These gradients would have provided a source of energy for driving active transport processes and potentially powering early metabolic reactions. Adaptations to environmental stressors, such as changes in temperature, pH, or salinity, would have been necessary for the survival of these early membrane-bound systems. This would have required the evolution of membrane compositions capable of maintaining integrity under varying conditions. The emergence of simple signaling mechanisms across these membranes would have been important for responding to environmental changes. This might have involved the development of primitive receptors or environmentally sensitive membrane components. Mechanisms for the controlled fusion and fission of these early membrane-bound compartments would have had to develop. This would have been crucial for the exchange of genetic material and other essential components between protocells, potentially facilitating early forms of horizontal gene transfer. The co-evolution of membrane permeability with internal metabolic processes would have been essential. As more complex chemical reactions developed within these compartments, the membrane's selective permeability would have had to adapt to support these processes, creating a feedback loop driving further complexity. These interconnected developments in membrane structure and function would have been fundamental in the transition from simple chemical systems to more complex, life-like entities capable of maintaining distinct internal environments and interacting with their surroundings in increasingly sophisticated ways.
Unresolved Issues and Conceptual Problems
2. Membrane-bound Enzyme Complexes
Acetoclastic methanogenesis relies on membrane-bound enzyme complexes, such as the CO dehydrogenase/acetyl-CoA synthase complex. This multi-subunit enzyme system is intricately integrated into the cell membrane, requiring specific lipid interactions and protein-protein associations.
Conceptual problem: Coordinated Assembly
- Lack of explanation for the spontaneous assembly of multi-subunit complexes
- Challenge in accounting for the precise spatial organization required for function
3. Energy Conservation Mechanisms
The pathway involves sophisticated energy conservation mechanisms, including the use of sodium ion gradients and the conversion of membrane potential to ATP via ATP synthase. The emergence of such intricate energy coupling systems poses significant challenges to unguided origin scenarios.
Conceptual problem: Energy Coupling Complexity
- No clear path for the emergence of chemiosmotic energy conservation
- Difficulty explaining the origin of ATP synthase's rotary mechanism
4. Cofactor Biosynthesis
Acetoclastic methanogenesis requires unique cofactors, such as coenzyme M and methanofuran. The biosynthetic pathways for these cofactors are complex and specific to methanogens.
Conceptual problem: Cofactor-Enzyme Interdependence
- Chicken-and-egg scenario: cofactors needed for enzymes, enzymes needed for cofactor synthesis
- No known prebiotic routes for complex cofactor synthesis
5. Methyl-Transfer Reactions
The pathway involves several methyl-transfer reactions, requiring specialized methyltransferases and methyl carriers. These reactions are highly specific and often involve unusual chemistry.
Conceptual problem: Chemical Novelty
- Difficulty explaining the origin of novel chemical mechanisms
- Challenge in accounting for the emergence of specific methyl carriers
6. Reverse Electron Transport
Acetoclastic methanogenesis employs reverse electron transport to generate reducing power. This process requires a precisely tuned electron transport chain and coupling mechanisms.
Conceptual problem: Thermodynamic Challenges
- No clear explanation for the emergence of energetically unfavorable electron transport
- Difficulty in accounting for the fine-tuning required for efficient energy conservation
7. Archaeal Membrane Composition
Methanogenic archaea possess unique membrane lipids, including isoprenoid-based lipids with ether linkages. These lipids are crucial for maintaining membrane integrity under extreme conditions.
Conceptual problem: Lipid Specificity
- No known prebiotic routes for archaeal lipid synthesis
- Challenge in explaining the emergence of domain-specific membrane compositions
8. Gene Regulation and Metabolic Control
Acetoclastic methanogenesis requires sophisticated gene regulation and metabolic control mechanisms to respond to environmental changes and substrate availability.
Conceptual problem: Regulatory Complexity
- Difficulty in explaining the origin of complex regulatory networks
- Challenge in accounting for the coordination of multiple metabolic pathways
9. Anaerobic Adaptations
Methanogens are obligate anaerobes with specific adaptations to low-redox environments. These adaptations include oxygen-sensitive enzymes and unique electron carriers.
Conceptual problem: Environmental Specialization
- No clear explanation for the emergence of highly specialized anaerobic metabolism
- Challenge in accounting for the development of oxygen sensitivity mechanisms
10. Methanogen-Specific Protein Families
Acetoclastic methanogens possess several protein families unique to their lineage, with no clear homologs in other organisms. The origin of these methanogen-specific proteins remains unexplained.
Conceptual problem: Protein Novelty
- Difficulty in explaining the emergence of entirely new protein families
- Challenge in accounting for the functional integration of novel proteins
These unresolved challenges highlight the significant gaps in our understanding of how acetoclastic methanogenesis could have emerged through unguided processes. The complexity, specificity, and interconnectedness of the various components involved in this metabolic pathway pose substantial conceptual problems for naturalistic explanations of its origin. Further research is needed to address these challenges and provide a comprehensive account of the emergence of this sophisticated biochemical system.
7.9.4 Overcoming Thermodynamic Barriers in Prebiotic Molecular Synthesis
Thermodynamic barriers would have had to be overcome through several key mechanisms to enable the formation of complex prebiotic molecules:
1. Coupling to exergonic reactions: Energy-releasing reactions would have driven thermodynamically unfavorable processes. For example, phosphodiester bond synthesis in nucleic acids would have been coupled with the breakdown of energy-rich compounds like pyrophosphates.
2. High-energy intermediates: Activated phosphate compounds, such as acetyl phosphate or phosphoenolpyruvate, would have facilitated energetically demanding steps in molecular synthesis, providing the necessary energy to form bonds that are difficult to create under prebiotic conditions.
3. Environmental energy sources:
a) Geothermal heat from hydrothermal vents could have driven endergonic reactions and created temperature gradients conducive to molecular concentration and synthesis.
b) UV radiation might have initiated photochemical reactions, forming high-energy precursors or driving bond formation in organic molecules.
c) Redox gradients, particularly in hydrothermal vent systems, could have provided a continuous source of chemical potential energy to drive unfavorable reactions.
4. Mineral surfaces would have acted as catalysts, lowering activation energies for key reactions and stabilizing reaction intermediates.
5. Concentration mechanisms, such as adsorption on mineral surfaces or evaporation cycles, would have increased local reactant concentrations, driving reactions forward despite unfavorable equilibrium constants.
6. Selective stabilization of products: This could have occurred through incorporation into larger molecular assemblies or binding to specific surfaces, shifting equilibria towards product formation.
7. Autocatalytic reaction networks: The emergence of systems where the products of certain reactions catalyze their own formation would have created self-amplifying processes capable of overcoming thermodynamic barriers.
These mechanisms, working together, would have been crucial in enabling the gradual accumulation of complex prebiotic molecules, setting the stage for the emergence of self-replicating systems and the origin of life, despite the unfavorable thermodynamics of many of these processes.
Challenges in Explaining Prebiotic Molecular Synthesis Without Guided Processes
1. Thermodynamic Hurdles in Biomolecule Formation:
The synthesis of complex biomolecules faces significant thermodynamic barriers that are difficult to overcome without guided processes.
a) Energy Requirements:
Many necessary reactions, such as peptide bond formation in proteins and phosphodiester bond formation in nucleic acids, are endergonic and require significant energy input.
Conceptual Problem: Energetic Implausibility
- No clear mechanism provides the required energy consistently in prebiotic environments.
- Difficulty explaining how endergonic reactions could have occurred spontaneously and repeatedly.
b) Concentration Dilemma:
Dilute prebiotic oceans posed significant challenges to molecular synthesis due to low reactant concentrations, but concentration mechanisms introduce other complications.
Conceptual Problem: Concentration Paradox
- Dilute solutions inhibit complex molecule formation.
- Concentration mechanisms like tidal pools introduce problems like hydrolysis and side reactions.
2. Chirality and Homochirality:
The emergence of homochirality in biological molecules presents a significant challenge.
a) Symmetry Breaking:
Abiotic processes usually produce racemic mixtures, yet life uses only one enantiomer for each type of chiral molecule, such as L-amino acids and D-sugars.
Conceptual Problem: Spontaneous Symmetry Breaking
- No known mechanism consistently produces enantiopure compounds abiotically.
- Difficulty explaining the origin of homochirality without invoking selective processes.
b) Amplification and Maintenance:
Even with a slight enantiomeric excess, maintaining and amplifying this excess over time remains problematic.
Conceptual Problem: Chiral Stability
- No clear mechanism for amplifying small enantiomeric excesses.
- Difficulty maintaining homochirality in the face of racemization processes.
3. Sequence Specificity in Informational Polymers:
The origin of sequence-specific polymers, which are crucial for information storage and catalysis, poses significant challenges.
a) Random vs. Functional Sequences:
Random polymerization would generate a vast array of non-functional sequences, yet life requires specific sequences for proteins and nucleic acids.
Conceptual Problem: Functional Improbability
- No known mechanism preferentially produces functional sequences.
- The vast sequence space makes random formation of useful polymers highly improbable.
b) Information Content:
The origin of the genetic code and the information it carries remains unexplained without invoking guided processes.
Conceptual Problem: Information Emergence
- No clear mechanism exists for the spontaneous generation of complex, meaningful information.
- Difficulty explaining the origin of the genetic code and its universality.
4. Cooperative Systems and Autocatalysis:
The emergence of cooperative systems and autocatalytic networks, which are critical for early metabolic processes, faces challenges.
a) Network Complexity:
Autocatalytic networks require multiple components to work in concert, raising questions about their spontaneous formation.
Conceptual Problem: Simultaneous Emergence
- No known mechanism explains the simultaneous emergence of multiple, interdependent components.
- Difficulty explaining how complex networks could arise without pre-existing templates.
b) Catalytic Efficiency:
Early catalysts were likely inefficient, raising questions about how they could have driven meaningful reactions.
Conceptual Problem: Catalytic Threshold
- No clear mechanism exists for improving catalytic efficiency without a selection process.
- Difficulty explaining how inefficient early catalysts could have sustained proto-metabolic networks.
5. Compartmentalization and Protocells:
The formation of protocells, necessary for creating distinct chemical environments, faces several hurdles.
a) Membrane Formation:
The spontaneous assembly of stable, semi-permeable membranes from prebiotic compounds is problematic.
Conceptual Problem: Membrane Stability
- No known mechanism consistently produces stable membranes from available prebiotic molecules.
- Difficulty explaining the origin of selective permeability without complex, evolved transport systems.
b) Encapsulation and Growth:
Explaining how protocellular structures encapsulated necessary components and grew/divided is challenging.
Conceptual Problem: Coordinated Assembly
- No clear mechanism exists for simultaneously encapsulating all necessary components for proto-life.
- Difficulty explaining coordinated growth and division without pre-existing regulatory systems.
6. Transition from Chemistry to Biology:
The transition from complex chemical systems to living entities presents perhaps the most significant challenge.
a) Self-Replication:
The emergence of true self-replication, as opposed to simple autocatalysis, remains unexplained.
Conceptual Problem: Replication Complexity
- No known mechanism explains the spontaneous emergence of accurate self-replication.
- Difficulty explaining the origin of the complex machinery required for DNA replication without invoking guided processes.
b) Metabolism-First vs. Replication-First:
Both major hypotheses for the origin of life—metabolism-first and replication-first—face significant challenges upon closer examination.
Conceptual Problem: Chicken-and-Egg Dilemma
- No clear mechanism exists for establishing complex metabolic networks without genetic information.
- Difficulty explaining the emergence of replication systems without pre-existing metabolic support.
These challenges highlight the conceptual problems faced when explaining the origin of life through purely unguided, naturalistic processes. From the formation of basic building blocks to the emergence of self-replicating systems, each step presents hurdles that current scientific understanding struggles to overcome without invoking some form of guidance or design. The complexity, specificity, and interdependence observed in even the simplest living systems raise profound questions about the adequacy of purely chance-based explanations for life’s origin.
Last edited by Otangelo on Tue Nov 12, 2024 7:21 pm; edited 6 times in total