LUCA's minimal Physiology and ProteomeReconstructing a supposed universal common ancestor (UCA) is a fascinating but challenging endeavor in the field of evolutionary biology. The UCA, often referred to as the "Last Universal Common Ancestor" (LUCA), is believed to be the hypothetical organism from which all life on Earth descended. While scientists have made significant progress in understanding the characteristics and possible traits of LUCA, there are numerous challenges and complexities involved in this endeavor.
Key Cellular Processes and Functions in LUCACellular Infrastructure and IntegrityMembrane Proteins, and Transport: Ensuring nutrient uptake, waste removal, and environmental sensing.
Peptidoglycan Synthesis: Key for structural integrity in some cells.
Families/functions involved in various aspects of cell division in LUCA: Fundamental for reproduction.
Energy and MetabolismEnergy Metabolism, Central Carbon Metabolism, and Other Specific Pathways: Driving cellular activities.
One-Carbon Metabolism: A central metabolic pathway.
Fatty Acid and Phospholipid Synthesis in LUCA: Critical for membrane structure and function.
Information Storage and ProcessingDNA Processing in LUCA: Replication, repair, and recombination.
Gene Expression and Regulation in LUCA: Transcribing DNA into functional RNA molecules.
Transcription/regulation in the LUCA: Ensuring correct gene expression.
Translation/Ribosome in the LUCA: Synthesizing proteins based on RNA templates.
Biosynthesis and Assembly of the Bacterial Ribosome: Critical for protein synthesis.
Biosynthesis and Nutrient UptakeAmino acid biosynthesis: Building blocks for proteins.
Nucleotide Synthesis and Salvage: Building blocks for DNA/RNA.
Polyamine Synthesis: Involved in DNA stabilization and protein synthesis.
Cofactor and Metal Cluster Biosynthesis: Essential for various enzyme functions.
Regulation and Response MechanismsEpigenetic, manufacturing, signaling, and regulatory codes in LUCA: Adapting to environmental changes and ensuring cellular homeostasis.
Reactive oxygen species (ROS): Handling oxidative stress.
Thermo protection in the LUCA: Protecting against temperature extremes.
Regulatory Enzymes and Proteins in Amino Acid Synthesis: Ensuring metabolic balance.
Proteolysis in the LUCA: Protein degradation and turnover.
Metals and Metal Clusters
Metal clusters: Essential for many enzyme functions.
RNA DynamicsRNA: Beyond protein-coding, RNA plays roles in regulation, catalysis, and structural components.
Additional FunctionsIon Homeostasis and Osmoregulation: Balancing ion concentrations and responding to osmotic stress.
DNA Repair Mechanisms: Maintaining genetic integrity in the face of damage.
Toxin-Antitoxin Systems: Protecting against foreign DNA or controlling cellular processes.
Sensory Systems: Detecting environmental signals and responding appropriately.
Vesicle Formation and Secretion: Interacting with the environment, disposing of waste, or communication.
There is no consensus viewThe absence of a consensus view on the constitution of the Last Universal Common Ancestor (LUCA) stems from several inherent challenges and uncertainties in the field of evolutionary biology and the study of ancient life forms:
Studies aiming to reconstruct the LUCA's genome or proteome are based on inference and computational analysis. They work backward in time from the characteristics of modern organisms to make educated guesses about the traits of LUCA. These inferences are reliant on various assumptions and models, which can introduce bias and errors into the predictions. LUCA is believed to have existed over 3.5 billion years ago. The vast amount of time that has passed since LUCA's existence makes it extremely challenging to accurately reconstruct its genetic and biochemical makeup. Over such a long period, genetic material can be lost, altered, or replaced. Gene families may have been present in LUCA but lost during subsequent evolution. Additionally, some functions may have been encoded by different, unrelated genes in LUCA and modern organisms due to gene replacement events. These factors can obscure the true nature of LUCA's genetic repertoire. The genomic data used in these studies come from modern organisms, and there may be gaps in our understanding of the diversity of ancient life. Some lineages that could have been present in the ancient microbial communities might have left no descendants or left only limited genomic traces. Each study on LUCA reconstruction employs its own set of methods, assumptions, and criteria for defining ancestral traits. There is no standardized or universally accepted protocol for these reconstructions, leading to variability in results. The available genomic and proteomic data used in these studies might not be representative of the full diversity of life that existed at the time of LUCA. The data could be biased toward organisms that are more easily studied or preserved. The concept of LUCA as a relatively complex cellular organism challenges traditional views of LUCA as a simple replicator. This complexity raises questions about the nature of the supposed LUCA and the extent to which it resembled modern organisms. Due to these challenges and uncertainties, different studies produce varying conclusions about LUCA's genome and proteome.
The vast diversity of life forms on Earth is an intriguing phenomenon. Instead of attributing it to a single common ancestor, an alternative hypothesis is that life forms were intentionally designed with unique characteristics and features. In this view, each species may have been individually created with its distinct traits. It is undeniable that all known life share a common biochemistry, including nucleic acids, proteins, carbohydrates, and lipids. Instead of seeing this as evidence of a LUCA, that might be due to a purposeful design. The shared biochemical foundation can be due to a blueprint or template used by an intelligent designer to create life forms with similar functional requirements. The complexity of life forms, including intricate cellular structures and metabolic pathways, can point to intelligent design. Instead of assuming that a simple replicator evolved into complex organisms, one can propose that these complexities were deliberately engineered.
Exploring what it would take for a transition from a LUCA to the three domains of life and beyond can help shed light on the plausibility of the tree of life hypothesis and whether evidence might rather suggest polyphyletic origins.
One of the significant challenges in the naturalistic hypothesis is, furthermore, explaining the origin of the intricate genetic information present in all life forms. From an alternative viewpoint, the sudden appearance of highly organized genetic codified information might raise questions about whether natural processes alone can account for the complexity of life's information systems. The transition from LUCA to the diverse life forms we observe today involves the development of complex molecular machinery within organisms. The instantiation of such sophisticated systems may indicate purposeful design or intervention, rather than a purely natural progression. The supposed evolution of cellular structures, organelles, and complex biochemical pathways required for life's functions presents significant challenges. The LUCA hypothesis implies that all life shares a common ancestry, but the vast differences in the developmental pathways and body plans of organisms deserve skepticism. The divergence in life forms' characteristics and features challenges the idea of a single common ancestor. Beyond the three domains of life, the evolution of consciousness, cognition, and complex behaviors in some organisms presents an additional layer of complexity. Examining the transition from LUCA to diverse life forms and beyond raises profound philosophical questions about the nature of existence, purpose, and the potential role of guiding intelligence in the universe. It encourages a broader discussion beyond purely scientific aspects.
LUCA was as complex as modern cellsIn 2005, the paper “A minimal estimate for the gene content of the last universal common ancestor—exobiology from a terrestrial perspective” by Christos A. Ouzounis et al aimed to reconstruct the gene content of the last universal common ancestor (LUCA), a hypothetical life form that presumably was the progenitor of the three domains of life. Using an algorithm for ancestral state inference of gene content, given a large number of extant genome sequences and a phylogenetic tree, they found that LUCA’s gene content appears to be substantially higher than that proposed previously, with a typical number of over 1000 gene families, of which more than 90% are also functionally characterized. More precisely, when only prokaryotes are considered, the number varies between 1006 and 1189 gene families while when eukaryotes are also included, this number increases to between 1344 and 1529 families depending on the underlying phylogenetic tree. Therefore, the common belief that the hypothetical genome of LUCA should resemble those of the smallest extant genomes of obligate parasites is not supported by recent advances in computational genomics. Instead, a fairly complex genome similar to those of free-living prokaryotes, with a variety of functional capabilities including metabolic transformation, information processing, membrane/transport proteins, and complex regulation, shared between the three domains of life, emerges as the most likely progenitor of life on Earth. 1F.Rana (2023): It now appears that LUCA was far more elaborate and complex than most evolutionary biologists had imagined. This conclusion comes from a team of collaborators from the US and the UK. In fact, the genetic complexity required to support LUCA’s morphological and physiological complexity far outpaces the estimated complexity from reconstructing LUCA based on comparative gene studies. In fact, the research team discovered that LUCA possessed a genome that was about 2.49 million base pairs in size (similar in size and complexity to a typical bacterial genome).2
The article: "Phenotypic reconstruction of the last universal common ancestor reveals a complex cell" discusses new research that offers a different perspective on LUCA and challenges previously held beliefs about its complexity. Most prior work focused on DNA to understand LUCA, but these studies might have underestimated its intricacy. The new approach taken by the international research team concentrates on LUCA's physiological features rather than its gene set, providing a more comprehensive view of its characteristics. From the information given: Historically, researchers thought LUCA was a simple cell. However, the new findings suggest that LUCA was perhaps more sophisticated than many contemporary bacteria and archaea. This challenges the idea that prokaryotic evolution was a journey from simple to complex. The study's abstract emphasized the rapid evolution of early life towards significant cellular complexity, much earlier than previously believed. Reconstructing LUCA's Features: Previously, genomics studies tried to deduce LUCA's gene set based on genes common to all bacteria and archaea. Estimates based on these methods suggested LUCA had around 350 to 1000 genes. But this approach had its limitations, particularly regarding understanding LUCA's morphology and molecular physiology. Instead of relying solely on DNA, the research team reconstructed LUCA's cellular traits using evolutionary trees constructed from 28 traits found in 3,128 bacterial and archaeal species. This methodology yielded a detailed set of morphological and physiological features for LUCA, painting a picture of a surprisingly complex organism. This method estimated that LUCA had a genome that was about
2.49 million base pairs in size, much larger than earlier estimates based on comparative gene studies. In essence, the article emphasizes that the traditionally accepted narrative of a simple LUCA, based on its supposed minimal gene content, may be incorrect. Instead, the new insights suggest that LUCA had a more complex gene content and was perhaps more intricate than previously thought. This discovery reshapes our understanding of early life's origin and suggests that life's complexity developed rapidly.
Kunin, V.,(2006): The Last Universal Common Ancestor (LUCA) or the primitive community that it encompassed was both functionally and genetically intricate. This supports the notion that life had reached its contemporary cellular structure well before the differentiation into the three domains. Contrary to conclusions drawn solely from sequence conservation and ubiquitous presence across all species, which pointed towards a basic LUCA with primarily translation-related genes, a refined approach to identify distant homology indicates that LUCA encompassed elements from almost all critical functional areas seen in current organisms. This reveals a metabolic complexity comparable to the translation process in terms of domain diversity. 1
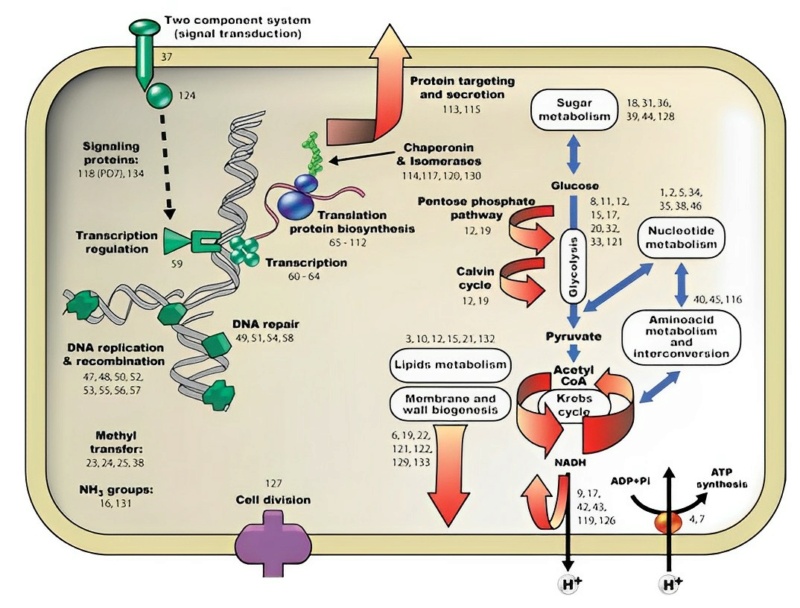
Schematic representation of cellular functions represented by the ancestral set of superfamilies. The cellular and/or functional locations of the superfamilies domains are represented by numbers. CATH identifications and functional descriptions of all ancestral superfamilies are given in Supplementary Table 3 following the same numbering code. 3
References● Kyrpides, N., Overbeek, R., & Ouzounis, C. (1999). Universal protein families and the functional content of the last universal common ancestor. Journal of Molecular Evolution, 49(4), 413-423. Link. (This study investigates the protein families universally present in living organisms, shedding light on the potential functional content of LUCA.)
●Mirkin BG, Fenner TI, Galperin MY, Koonin EV (2003). Algorithms for computing parsimonious evolutionary scenarios for genome evolution, the last universal common ancestor and dominance of horizontal gene transfer in the evolution of prokaryotes. BMC Evol Biol, 3:2. Link. (This paper discusses algorithms for understanding genome evolution, the last universal common ancestor, and the role of horizontal gene transfer in prokaryotic evolution.)
●Yang S, Doolittle RF, Bourne PE (2005). Phylogeny determined by protein domain content. Proc Nat Acad Sci U S A, 102:373-378. Link. (The study explores how protein domain content can be used to determine phylogenetic relationships.)
●Delaye L, Becerra A, Lazcano A (2005). The last common ancestor: what's in a name?. Orig Life Evol Biosph, 35:537-554. Link. (This paper delves into the terminological and conceptual challenges associated with defining the last common ancestor.)
●Ranea JA, Sillero A, Thornton JM, Orengo CA (2006). Protein superfamily evolution and the last universal common ancestor (LUCA). J Mol Evol, 63:513–525. Link. (The paper studies the evolution of protein superfamilies with reference to the last universal common ancestor.)
●Wang M, Yafremava LS, Caetano-Anollés D, Mittenthal JE, Caetano-Anollés G (2007). Reductive evolution of architectural repertoires in proteomes and the birth of the tripartite world. Genome Res, 17:1572-1585. Link. (This paper discusses the reductive evolution in proteomes and how it may have given rise to the tripartite division of life forms.)
●Srinvasan V, Morowitz HJ (2009). The canonical network of autotrophic intermediary metabolism: minimal metabolome of a reductive chemoautotroph. Biol Bull, 216:126–130. Link. (This paper describes the minimal metabolic network required for a reductive chemoautotroph.)
● Goldman AD, Baross JA, Samudrala R. (2012) The Enzymatic and Metabolic Capabilities of Early Life. PLoS ONE. Link. (This study investigates the enzymatic and metabolic attributes of early life forms, offering insights into primordial biochemistry.)
● de Farias, S. T., Rêgo, T. G., & José, M. V. (2015). A proposal of the proteome before the last universal common ancestor (LUCA). Published online by Cambridge University Press. Link. (This article offers a proposed view of the proteome that existed before the emergence of the Last Universal Common Ancestor, LUCA, providing insights into the evolutionary stages of protein networks in primordial life forms.)
●Weiss MC, Sousa FL, Mrnjavac N, Neukirchen S, Roettger M, Nelson-Sathi S, Martin WF (2016). The physiology and habitat of the last universal common ancestor. Nat Microbiol, 1:16116. Link. (This paper explores the potential physiology and habitat of the last universal common ancestor based on extant biochemistry and cell biology.)
●Harris A, Goldman AD (2018). Phylogenetic Reconstruction Shows Independent Evolutionary Origins of Mitochondrial Transcription Factors from an Ancient Family of RNA Methyltransferase Proteins. J Mol Evol, 86:277-282. Link. (The paper focuses on the independent evolutionary origins of mitochondrial transcription factors.)
● Palacios-Pérez, M., & José, M. V. (2019). The evolution of proteome: From the primeval to the very dawn of LUCA. BioSystems. Link. (This research article explores the evolution of the proteome from the earliest stages of life up to the emergence of the Last Universal Common Ancestor, LUCA, providing insights into the evolution of proteins and protein networks in early life forms.)
● Crapitto, A. J., Campbell, A., Harris, A.J., & Goldman, A. D. (2022). A consensus view of the proteome of the last universal common ancestor. Ecology and Evolution, 12(6), e8930. Link. (Published by Wiley, this article presents a consensus perspective on the proteome of the Last Universal Common Ancestor, LUCA. The study contributes to the field of biology and provides insights into the protein composition of early life forms.)
● Matzke, N., Lin, A., Stone, M., & Baker, M. (2021). Flagellar export apparatus and ATP synthetase: Homology evidenced by synteny predating the Last Universal Common Ancestor. BioEssays, 43. Link. (Published by Wiley, this article delves into the homological relationship between the flagellar export apparatus and ATP synthetase, emphasizing their ancestral connection predating the Last Universal Common Ancestor. This work enhances our understanding of evolutionary biology and the genetic underpinnings of ancient cellular machinery.)
Note: This comprehensive list captures both known functions and enzymes related to DNA processing in LUCA as well as recent additions based on updated knowledge. The evolutionary history and precise functions of these enzymes might vary, and our understanding remains a subject of active research.
1.Ouzounis, C. A., Kunin, V., Darzentas, N., & Goldovsky, L. (2006). A minimal estimate for the gene content of the last universal common ancestor--exobiology from a terrestrial perspective. Research in Microbiology, 157(1), 57-68. Link. (This study provides an estimate of the gene content of the Last Universal Common Ancestor, offering insights into its molecular features and potential exobiological implications.)2.Rana, F. (2023).
LUCA’s Complexity Challenges Evolutionary Origin of Life. The Cell's Design Blog. Link. (This article delves into the newfound complexity of LUCA and the challenges it presents to the evolutionary model for the origin of life.)
Metabolic Pathways in LUCAUnderstanding which metabolic pathways were extant in LUCA is an ongoing area of scientific research. The listed pathways come from interpretations of comparative genomics studies, phylogenetic analyses, and biochemical studies. However, as with many topics in ancient evolution, there's a degree of uncertainty and some pathways may be subject to revision as more data becomes available. Here's a list of potential metabolic pathways that might have been present in LUCA, keeping in mind that this is based on our current understanding and can change with future discoveries:
Anabolic Pathways (Biosynthetic)1. Nucleotide Synthesis and Salvage: The formation of the basic units of nucleic acids lays the groundwork for genetic information storage and transfer.
2. Amino Acid Biosynthesis: Proteins serve as the primary workhorses of the cell, and their constituent amino acids would be among the earliest necessities.
3. Fatty Acid and Phospholipid Synthesis: Once basic biochemical pathways were established, compartmentalization via cellular membranes would be crucial for the emergence of protocells and differentiation from the environment.
4. One-Carbon Metabolism: Foundational biochemical reactions that likely evolved early, supporting the synthesis of various vital molecules.
5. RNA Modifications: RNA's role as both a genetic material and catalytic molecule would necessitate modifications for stability and function.
6. Peptidoglycan Synthesis: With the emergence of a lipid barrier, a protective layer or primitive cell wall would be beneficial, especially for early bacterial life.
7. Coenzyme A and Other Cofactor Biosynthesis: As metabolic reactions diversified, molecules that aid in enzyme function and catalysis would become indispensable.
8. Vitamin and Cofactor Biosynthesis: Further diversification of enzymatic reactions would drive the biosynthesis of more specialized cofactors and vitamins.
9. Iron-Sulfur Cluster Assembly: Reflecting an early Earth rich in iron and underscoring the significance of iron-sulfur proteins in primordial metabolic processes.
10. Porphyrin Metabolism: As life evolved, processes like respiration and photosynthesis would emerge, necessitating molecules like heme and chlorophyll.
11. Polyamine Synthesis: With increased cellular complexity, polyamines would be essential for stabilizing DNA, RNA, and supporting cell growth.
Catabolic Pathways (Degradative)12. Glycolysis and the Pentose Phosphate Pathway: Fundamental pathways for sugar breakdown and nucleotide synthesis.
13. TCA Cycle: Core pathway for cellular breathing, energy production, and precursors for biosynthesis.
14. ATP Synthesis and Oxidative Phosphorylation: Key processes for generating ATP, the main energy currency of cells.
15. Anaerobic Respiration Pathways: Pathways that allow energy production without oxygen.
16. Methanogenesis and Methane Oxidation: Enzymatic evidence suggests methane metabolism in early evolutionary pathways.
17. Hydrogen Oxidation: Pathways for energy derivation from hydrogen.
18. Thioester Metabolism: Points to thioesters as ancient energy carriers.
19. Carbon Fixation: Mechanisms for assimilating CO2 into organic molecules.
20. Methyl Group Metabolism: Related to the vital transfer and use of methyl groups in cellular functions.
21. Reverse Electron Flow: Used for driving thermodynamically challenging reactions.
22. Formate Metabolism: Formate as a substrate and product in early life energy production.
23. Entner–Doudoroff pathway: An alternate to glycolysis, crucial for glucose breakdown.
24. Phosphoketolase pathway: Focused on pentose sugars, primarily in prokaryotes.
25. Wood-Ljungdahl Pathway: One of the earliest CO2 fixation pathways, suggesting autotrophic origins of life.
26. Acetogenesis: Production of acetate from CO2 and H2, which might have played a role in early energy metabolism.
Both Anabolic and Catabolic (as they serve dual purposes in the cell)27. Central Carbon Metabolism: Including the glycolytic pathway, pentose phosphate pathway, and the TCA cycle, these pathways are fundamental for energy production and the synthesis of precursors for various biomolecules.
28. Lipid Metabolism: Pathways focused on the synthesis, modification, and breakdown of lipids.
29. Nucleotide Degradation and Salvage Pathways: Crucial for recycling and recovering nucleotides.
30. Amino Acid Degradation: Breakdown of amino acids for energy production or conversion to other molecules.
31. Autophagy: The cellular process of breaking down and recycling cellular components.
32. Protein Degradation: Processes for breaking down and recycling proteins.
33. Lysosome Function: Involving the breakdown of cellular materials.
34. DNA Repair Mechanisms: Potential early systems to maintain genetic integrity.
 Ancestral enzyme functions determined from consensus LUCA
Ancestral enzyme functions determined from consensus LUCA eggNOG clusters mapped onto a universal metabolic network. The consensus LUCA enzyme functions are represented by 169 Enzyme Commission codes. The universal metabolic network and color-coding of metabolic categories are from the global Metabolic Pathways network (map 01100) from the KEGG database (Kanehisa et al., 2017; Ogata et al., 1999). “Metabolism of Other Amino Acids” is terminology that the KEGG database uses to indicate amino acids that are not included in proteins, such as D-amino acids
References● Srinivasan V, Morowitz HJ (2009). The canonical network of autotrophic intermediary metabolism: minimal metabolome of a reductive chemoautotroph. Biol Bull, 216:126–130. Link. (This paper describes the minimal metabolic network required for a reductive chemoautotroph, offering insights into LUCA's potential metabolic pathways.)
● Weiss MC, Sousa FL, Mrnjavac N, Neukirchen S, Roettger M, Nelson-Sathi S, Martin WF (2016). The physiology and habitat of the last universal common ancestor. Nat Microbiol, 1:16116. Link. (This paper explores the potential physiology and habitat of LUCA, delving into its possible metabolic mechanisms based on extant biochemistry and cell biology.)
● Wimmer, J. L. E., Xavier, J. C., Vieira, A. d. N., Pereira, D. P. H., Leidner, J., Sousa, F. L., Kleinermanns, K., Preiner, M., & Martin, W. F. (2021). Energy at Origins: Favorable Thermodynamics of Biosynthetic Reactions in the Last Universal Common Ancestor (LUCA). Front. Microbiol., 12. Link. (This paper is part of a research topic on the Last Universal Common Ancestor and Origin of Life. It explores the favorable thermodynamics of biosynthetic reactions in the LUCA, shedding light on the energy considerations at the origin of life.)




