15. DNA Methylation
DNA methylation is an epigenetic modification that involves the addition of a methyl group to the cytosine base of a DNA molecule, typically occurring at cytosine-guanine dinucleotide sequences (CpG sites). This modification is catalyzed by enzymes called DNA methyltransferases. DNA methylation plays a crucial role in regulating gene expression by modulating the accessibility of DNA to transcription factors and other regulatory proteins. In DNA methylation, a methyl group (CH3) is added to the carbon-5 position of the cytosine ring. This modification can lead to changes in chromatin structure, making the associated DNA regions more condensed and less accessible for transcriptional machinery. As a result, genes in methylated regions are often silenced or have reduced transcriptional activity. Conversely, unmethylated regions are generally associated with active gene expression.
Importance in Biological Systems
DNA methylation is essential for various biological processes and cellular functions:
Gene Regulation: DNA methylation plays a central role in controlling gene expression. Methylated DNA segments prevent the binding of transcription factors and other regulatory proteins, inhibiting gene transcription.
Cellular Identity and Differentiation: DNA methylation patterns are established during development and are crucial for determining cell identity. They contribute to the differentiation of cells into various specialized types in multicellular organisms.
X-Chromosome Inactivation: In females, DNA methylation is involved in X-chromosome inactivation, a process that compensates for the presence of two X chromosomes. One X chromosome in each cell becomes highly methylated and largely inactive.
Epigenetic Memory: DNA methylation patterns can be heritably passed on to daughter cells during cell division. This epigenetic memory contributes to maintaining cell identity and lineage-specific gene expression.
Tumor Suppression: DNA methylation can act as a tumor suppressor mechanism by silencing genes involved in cell cycle regulation and DNA repair. Aberrant DNA methylation is associated with cancer development.
Developmental Processes Shaping Organismal Form and Function
DNA methylation is particularly important in shaping organismal form and function during developmental processes:
Embryonic Development: DNA methylation patterns undergo dynamic changes during embryonic development. They guide the differentiation of cells into distinct lineages, leading to the formation of tissues and organs.
Tissue-Specific Gene Expression: DNA methylation contributes to establishing tissue-specific gene expression profiles. Different cell types exhibit unique methylation patterns that help maintain their specialized functions.
Imprint Control: DNA methylation controls imprinted genes, which are expressed in a parent-of-origin-specific manner. Imprinting influences aspects of growth, development, and behavior.
Adaptation to Environment: DNA methylation can respond to environmental cues and influence gene expression accordingly. This process allows organisms to adapt to changing environmental conditions.
DNA methylation is a fundamental epigenetic modification that plays a pivotal role in gene regulation, cellular differentiation, developmental processes, and the establishment of cell identity. Its dynamic and context-dependent nature makes it a critical factor in shaping the form and function of organisms throughout their life cycles.
What is the significance of DNA methylation in regulating gene expression and cell differentiation during development?
DNA methylation plays a significant role in regulating gene expression and cell differentiation during development by influencing the accessibility of genes to the transcriptional machinery and guiding cells toward specific lineages. Here's how DNA methylation contributes to these processes:
Gene Expression Regulation
Silencing Gene Expression: DNA methylation at promoter regions of genes can physically block the binding of transcription factors and other regulatory proteins. This prevents the initiation of transcription, effectively silencing gene expression.
Stable Repression: DNA methylation provides a stable and heritable form of gene repression. Once established, methylation patterns can be maintained through DNA replication and cell division, ensuring long-term control over gene expression.
Tissue-Specific Expression: DNA methylation patterns are often tissue-specific. By methylating certain genes in specific cell types, the expression of those genes can be restricted to certain tissues, contributing to the development of distinct cell lineages.
Cell Differentiation and Development
Epigenetic Memory: DNA methylation patterns acquired during early development can serve as epigenetic memory, ensuring that cells maintain their specialized functions as they divide and differentiate. This contributes to the stability of cell identities.
Lineage Commitment: DNA methylation patterns guide cells toward specific lineages during differentiation. By silencing genes associated with alternative cell fates, DNA methylation helps commit cells to their intended developmental pathway.
Imprinting and Parental Alleles: DNA methylation is involved in genomic imprinting, where certain genes are expressed based on their parental origin. Imprinting is critical for proper development, as it ensures the appropriate expression of genes that influence growth and metabolism.
X-Chromosome Inactivation: DNA methylation is responsible for X-chromosome inactivation in females. It ensures that one of the two X chromosomes in each cell is largely silenced, preventing an imbalance in gene dosage between males and females.
Cell Identity Maintenance: DNA methylation helps maintain cell identity by ensuring that only relevant genes are expressed in a given cell type. Changes in DNA methylation patterns can disrupt this identity and lead to aberrant cell behavior.
Adaptation and Plasticity
Environmental Response: DNA methylation can be influenced by environmental factors, allowing organisms to adapt to changing conditions. Changes in methylation patterns can modulate gene expression in response to environmental cues.
Developmental Plasticity: During early development, DNA methylation can confer a degree of plasticity, allowing cells to adopt different fates under specific conditions. As development progresses, methylation patterns become more stable and guide cellular differentiation.
How are DNA methylation patterns established and maintained through cell divisions?
The establishment and maintenance of DNA methylation patterns through cell divisions involve a combination of enzymatic processes, DNA replication, and epigenetic memory mechanisms. These processes ensure the faithful transmission of epigenetic information from one generation of cells to the next. Here's how DNA methylation patterns are established and preserved:
Establishment of DNA Methylation Patterns
De Novo Methylation: During early embryonic development, de novo DNA methylation occurs, where specific regions of the genome are marked with methyl groups. This process is catalyzed by enzymes called DNA methyltransferases (DNMTs).
Maintenance Methylation: Maintenance methylation refers to the replication-dependent addition of methyl groups to the newly synthesized DNA strand during DNA replication. The enzyme DNMT1 plays a crucial role in this process. DNMT1 recognizes hemi-methylated CpG sites (one methylated strand and one unmethylated strand) and adds a methyl group to the newly synthesized strand, thereby maintaining the methylation pattern after DNA replication.
Maintenance of DNA Methylation Patterns Through Cell Divisions
Epigenetic Memory: DNA methylation patterns can be maintained across multiple cell divisions due to epigenetic memory mechanisms. These mechanisms involve interactions between DNMTs and histone modifications that help stabilize the methylation pattern.
DNMT1 and Replication: As mentioned earlier, DNMT1 plays a key role in maintaining DNA methylation patterns during cell division. It ensures that the newly synthesized DNA strand is methylated based on the pattern of the template strand. This process helps preserve the original methylation pattern in daughter cells.
Histone Modifications: Histone modifications and DNA methylation are closely linked. Certain histone modifications contribute to the recruitment of DNMTs to specific genomic regions. This coordination helps ensure that DNA methylation is faithfully maintained during replication and cell divisions.
Passive Demethylation Prevention: Passive demethylation occurs when methylated cytosines are not maintained during DNA replication. Histone modifications and DNMT1 activity prevent passive demethylation by ensuring that newly synthesized DNA strands are promptly methylated.
Epigenetic Plasticity and Adaptation
Environmental Influence: While DNA methylation patterns are generally stable, certain environmental factors can influence them. Environmental cues can lead to changes in DNA methylation patterns, allowing cells to adapt to new conditions and influences.
Epigenetic Variability: Despite the stability of DNA methylation, some degree of epigenetic variability exists between individuals and cell types. This variability can contribute to phenotypic diversity and may have implications for health and disease.
DNA methylation patterns are established during early development through de novo methylation and are then maintained through cell divisions via processes like maintenance methylation. Epigenetic memory mechanisms, interactions between DNMTs and histone modifications, and the faithful action of DNMT1 play crucial roles in preserving the methylation pattern. This stability contributes to the maintenance of cell identity, gene expression patterns, and proper development throughout the life of an organism.
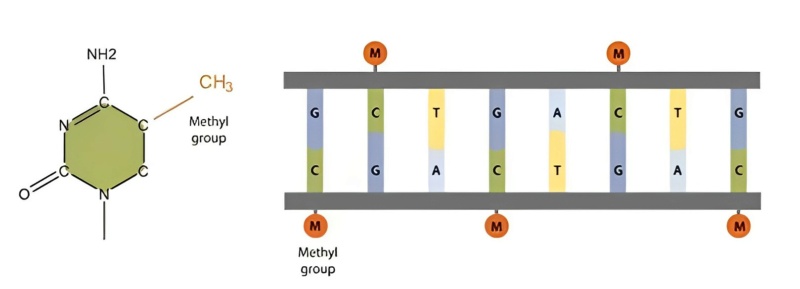
DNA methylation plays a pivotal role in the epigenetic regulation of gene expression. Specifically, the addition of a methyl group to the carbon 5 position of cytosine within cytosine-phosphate-guanine (CpG) and other nucleotide sequences has profound effects on gene activity. This epigenetic modification acts as a molecular "off" switch, inhibiting the binding of transcription factors to gene promoters. This simple chemical alteration introduces a layer of complexity to the genome's regulatory landscape, influencing how genes are read and interpreted by the cellular machinery. By methylating CpG sites in promoter regions, DNA methylation interferes with the recruitment of transcription factors—proteins that play a key role in initiating gene transcription. The binding of transcription factors to promoter regions is a crucial step in the process of gene activation. DNA methylation at these sites essentially creates a physical barrier that hinders the transcription factors from binding effectively, leading to reduced or silenced gene expression. This mechanism of DNA methylation-induced gene silencing has far-reaching implications for cellular processes, development, and health. It enables cells to fine-tune gene expression patterns, allowing them to respond to environmental cues, differentiate into specialized cell types, and maintain epigenetic memory. The intricate orchestration of DNA methylation and its interactions with other epigenetic marks, chromatin structure, and transcriptional machinery illustrate the elegant complexity of gene regulation within the cell. This interplay underscores the sophisticated control mechanisms that exist to ensure the appropriate functioning of our genetic material.
Appearance of DNA Methylation in the evolutionary timeline
The appearance of DNA methylation in the evolutionary timeline is a complex topic that involves speculation and ongoing research. While the exact timing and sequence of events are not fully understood, here is a general overview of the hypothesized appearance of DNA methylation in the evolutionary timeline:
Early Life Forms: DNA methylation is not likely to have played a significant role in early cellular life forms, as they lacked the organized cellular structures and regulatory mechanisms present in modern organisms.
Emergence of Prokaryotes: DNA methylation is hypothesized to have emerged relatively late in the evolution of life. Initial forms of DNA methylation would have been simple and limited in scope.
In prokaryotes, DNA methylation could have potentially played a role in the regulation of gene expression, although the extent and functions are not well established.
Transition to Eukaryotes: The appearance of eukaryotic cells would have brought about more complex regulatory mechanisms, including epigenetic modifications like DNA methylation. DNA methylation would have gained importance in eukaryotes as a mechanism for gene regulation, controlling processes such as cell differentiation and development.
Evolution of Multicellularity: With the emergence of multicellular organisms, DNA methylation would have become more intricate and diverse in its functions. The regulation of cell differentiation, tissue-specific gene expression, and the establishment of cell identities would have become crucial, making DNA methylation a key player in these processes.
Vertebrate Evolution and Complexity: Vertebrates, including mammals, exhibit highly complex DNA methylation patterns associated with various cellular processes. In vertebrates, DNA methylation plays roles in genomic imprinting, X-chromosome inactivation, and the regulation of tissue-specific gene expression.
Expansion of Functions and Epigenetic Landscape: Over time, DNA methylation would have evolved to have broader functions, influencing not only gene expression but also genome stability, response to environmental cues, and adaptation. The epigenetic landscape would have become more complex as additional enzymes and regulatory mechanisms evolved to modulate DNA methylation patterns.
It's important to note that the timeline presented here is speculative, and the actual appearance and evolution of DNA methylation may have occurred differently. The study of DNA methylation's evolutionary history is an active area of research, and ongoing discoveries continue to shape our understanding of its origin and function across different species.
De Novo Genetic Information necessary to instantiate DNA Methylation
Creating the mechanisms of DNA methylation de novo involves the generation and integration of new genetic information to establish the enzymes, regulatory elements, and recognition systems required for DNA methylation. Here's a simplified overview of the hypothetical process:
Generation of Enzymes: New genetic information would need to code for enzymes known as DNA methyltransferases (DNMTs). These enzymes recognize specific DNA sequences and catalyze the addition of methyl groups to cytosine bases.
Regulatory Elements and Promoters: Regulatory elements, including promoters and enhancers, would need to emerge to control the expression of DNMT genes. These elements ensure that DNMTs are produced at the right time and in the right cells.
Target Recognition Sequences: New genetic information would need to provide the sequences that DNMTs recognize as their target sites for methylation (CpG sites). These sequences would need to be distributed appropriately throughout the genome.
Accessory Proteins: The genetic information would also need to encode accessory proteins that interact with DNMTs, ensuring proper enzymatic activity, binding, and recruitment to target sites.
Epigenetic Memory Mechanisms: To maintain DNA methylation patterns during cell divisions, epigenetic memory mechanisms would need to emerge. This could involve interactions between DNA methylation and histone modifications or the development of specialized proteins that facilitate maintenance.
Establishing Methylation Patterns: The genetic information would need to specify the initial methylation patterns, indicating which regions of the genome should be methylated and which should remain unmethylated.
Integration of Regulatory Networks: New genetic information would need to establish regulatory networks that link DNA methylation to other cellular processes, such as transcription, DNA repair, and chromatin remodeling.
Integration with Replication Machinery: The genetic information would need to coordinate with the DNA replication machinery to ensure that newly synthesized DNA strands are methylated in a manner consistent with the original pattern.
Environmental Sensitivity: If the hypothetical system is designed to respond to environmental cues, additional genetic information would be needed to link DNA methylation patterns to specific environmental signals.
Integration into the Genome: The new genetic information would need to integrate seamlessly into the existing genome to ensure that the mechanisms of DNA methylation function harmoniously with other cellular processes.
This hypothetical process involves the orchestrated creation of genes, regulatory elements, sequences, and mechanisms that work together to establish and maintain DNA methylation.
It's important to note that this description simplifies a complex process and doesn't account for the intricate interactions and regulatory networks that would be required for the functional instantiation of DNA methylation.
Manufacturing codes and languages that would have to emerge and be employed to instantiate DNA Methylation
To transition from an organism without DNA methylation to one with a fully developed DNA methylation system, a complex set of manufacturing codes and languages would need to be created, instantiated, and orchestrated. These non-genetic components are integral for the establishment, regulation, and maintenance of the DNA methylation machinery. Here's an explanation of the manufacturing codes and languages involved in this process:
Enzymatic Codes and Protein Assembly: Generation of codes that dictate the precise sequence and structure of DNA methyltransferase enzymes (DNMTs). Creation of molecular chaperones that aid in the correct folding and assembly of DNMTs, ensuring their functional three-dimensional structure.
Regulatory Elements and Promoter Codes: Establishment of codes that define the regulatory elements controlling the expression of genes encoding DNMTs. Development of promoter codes that determine the strength, timing, and tissue-specificity of DNMT gene expression.
CpG Recognition Codes: Creation of codes that specify the recognition sequences for DNMTs to identify and methylate CpG sites in DNA. Design of binding motifs within DNMTs that enable accurate recognition and binding to CpG-rich regions.
Epigenetic Inheritance Codes: Generation of codes that enable the transmission of DNA methylation patterns from parent to daughter cells during DNA replication. Implementation of feedback loops and stability codes to prevent random changes in methylation patterns.
Interplay and Integration Codes: Development of codes that facilitate crosstalk between DNA methylation and other epigenetic modifications, such as histone modifications. Creation of codes that enable coordinated regulation of gene expression through the interplay of DNA methylation and chromatin structure.
Environmental Responsiveness Mechanisms (Optional): Design of codes that allow the DNA methylation system to respond to environmental cues, leading to dynamic changes in methylation patterns. Implementation of decoding mechanisms that interpret external signals and trigger appropriate adjustments in DNA methylation.
Feedback Control Languages: Establishment of languages that enable DNMTs and associated proteins to communicate feedback signals regarding methylation levels and patterns. Development of codes that regulate the activity of DNMTs based on the cellular context and methylation status.
Integration and Organization Codes: Creation of codes that ensure the proper integration of the DNA methylation machinery into the cellular context. Design of organizational codes that guide the spatial arrangement and functional interactions of DNMTs and related components. The orchestrated creation and integration of these manufacturing codes and languages are essential to establish a functional DNA methylation system.
This intricate interplay among diverse components highlights the complexity of the process and raises questions about the simultaneous emergence and coordination of these codes. Proponents of intelligent design argue that the presence of such sophisticated and interdependent systems supports the notion of purposeful design rather than a gradual, step-by-step evolutionary process.
Epigenetic Regulatory Mechanisms necessary to be instantiated to create DNA Methylation
The development of DNA methylation from scratch would require the creation and subsequent utilization of intricate epigenetic regulatory systems. These systems would work in tandem to establish, regulate, and maintain DNA methylation. Here's an overview of the epigenetic regulation and the collaborative systems involved:
Epigenetic Regulatory Systems
DNA Methyltransferases (DNMTs) and Regulatory Elements: Creation of regulatory elements that control the expression of genes encoding DNMTs. Design of mechanisms that ensure DNMT expression is fine-tuned based on developmental stages and cellular contexts.
Methylation Target Recognition and Binding: Generation of mechanisms that enable DNMTs to recognize specific DNA sequences (CpG sites) for methylation. Establishment of codes that ensure accurate binding and proper positioning of DNMTs at target sites.
Epigenetic Memory and Maintenance: Design of epigenetic memory systems that retain DNA methylation patterns during cell divisions. Integration of mechanisms that coordinate DNA methylation maintenance with DNA replication and chromatin remodeling.
Integration with Chromatin Modifications: Creation of codes that allow crosstalk and coordination between DNA methylation and histone modifications. Development of molecular mechanisms that modulate chromatin structure and accessibility in response to DNA methylation changes.
Collaborative Systems
Chromatin Remodeling Complexes: Collaborative interaction with chromatin remodeling complexes to expose target regions for methylation and facilitate proper DNMT binding.
Histone Modification Systems: Joint operation with histone modification systems to create an epigenetic landscape that guides DNA methylation to specific genomic regions.
DNA Repair and Replication Machinery: Collaborative coordination with DNA repair and replication machinery to ensure accurate maintenance of DNA methylation patterns during cell divisions.
Transcriptional Regulatory Networks: Collaboration with transcriptional regulatory networks to influence gene expression patterns through the modulation of DNA methylation status.
Environmental Sensing and Adaptation (Optional): Joint functioning with environmental sensing mechanisms to allow for dynamic changes in DNA methylation patterns in response to external cues.
Cell Signaling Pathways: Collaboration with cell signaling pathways to integrate cellular signals that influence DNA methylation patterns based on physiological conditions.
Epigenetic Enzyme Networks: Collaborative interaction with other epigenetic enzymes, such as histone modifiers and chromatin remodelers, to orchestrate coordinated changes in chromatin structure.
The successful establishment of DNA methylation would require an intricate interplay among these epigenetic regulatory systems and collaborative mechanisms. Each component must be precisely created and integrated to ensure accurate DNA methylation patterns, stable epigenetic memory, and proper functioning within the cellular context. The complexity of these interdependent systems raises questions about their simultaneous emergence and coordinated operation, which is evidence for the purposeful and coordinated origin of such complex cellular processes.
Signaling Pathways necessary to create, and maintain DNA Methylation
The emergence of DNA methylation from scratch would necessitate the creation and subsequent involvement of signaling pathways that orchestrate and regulate this epigenetic modification. These signaling pathways would be interconnected, interdependent, and engage in crosstalk with each other and other biological systems to ensure accurate DNA methylation patterns. Here's an overview of the hypothetical signaling pathways and their interactions:
Developmental Signaling Pathways
Creation of signaling pathways that respond to developmental cues and guide the establishment of DNA methylation patterns in specific cells and tissues. Interdependence with transcription factors and chromatin modifiers to coordinate gene expression and epigenetic regulation.
Environmental Sensing Pathways: Generation of pathways that sense environmental cues and enable dynamic changes in DNA methylation patterns in response to external conditions. Interconnection with cellular stress responses and adaptive mechanisms to optimize cellular function.
DNA Damage and Repair Signaling: Establishment of signaling pathways that detect DNA damage and trigger DNA repair mechanisms. Crosstalk with DNA methylation maintenance systems to ensure accurate restoration of methylation patterns after repair.
Cell Signaling Cascades: Creation of cascades that transduce extracellular signals, such as growth factors or hormones, into intracellular responses that influence DNA methylation. Interactions with transcriptional regulators and chromatin modifiers to modulate gene expression and epigenetic states.
Epigenetic Cross-Talk Pathways: Generation of pathways that facilitate communication between different epigenetic modifications, such as DNA methylation and histone modifications. Interplay with chromatin remodeling complexes to establish coordinated changes in chromatin structure.
Replication and Chromatin Dynamics Pathways: Development of pathways that synchronize DNA methylation maintenance with DNA replication and chromatin dynamics. Coordination with histone modification pathways to ensure proper chromatin packaging and accessibility.
Transcriptional Feedback Loops: Establishment of feedback loops that link DNA methylation status with gene expression levels. Interdependence with transcription factors and RNA processing pathways to fine-tune gene regulation.
Cellular Stress and Homeostasis Pathways: Creation of pathways that monitor cellular stress and maintain homeostasis by adjusting DNA methylation patterns. Crosstalk with cell cycle checkpoints and metabolic pathways to ensure cellular integrity.
These signaling pathways would interact extensively with each other and with broader biological systems to ensure the accurate establishment and maintenance of DNA methylation patterns. The complexity and interconnectedness of these pathways underscore the intricate regulatory networks that would need to be simultaneously created and coordinated to support the emergence of DNA methylation. Proponents of intelligent design argue that the orchestrated integration of these pathways points toward a purposeful and intentional design rather than a stepwise evolutionary process.
Regulatory codes necessary for maintenance and operation of DNA Methylation
The maintenance and operation of DNA methylation would require the instantiation and subsequent involvement of intricate regulatory codes and languages that govern various aspects of this epigenetic modification. These codes and languages play a crucial role in ensuring the stability, accuracy, and responsiveness of DNA methylation patterns. Here's an overview of the regulatory codes and languages involved:
Maintenance and Inheritance Codes: Establishment of codes that facilitate the faithful transmission of DNA methylation patterns to daughter cells during DNA replication. Development of mechanisms that ensure the maintenance of established methylation patterns through cell divisions.
Methylation Stability Codes: Creation of codes that prevent random changes in DNA methylation patterns, ensuring stability and epigenetic memory. Implementation of feedback loops that monitor and rectify deviations from the desired methylation state.
Context-Dependent Codes: Generation of codes that allow for context-dependent DNA methylation, meaning that patterns can be adapted to specific cellular environments or developmental stages. Incorporation of regulatory elements that respond to cell type-specific cues to establish appropriate methylation patterns.
Environmental Responsiveness Codes (Optional): Design of codes that enable DNA methylation patterns to be modulated in response to external environmental signals. Integration of regulatory networks that interpret environmental cues and trigger changes in DNA methylation status.
Crosstalk and Communication Codes: Establishment of codes that facilitate communication between DNA methylation machinery and other cellular processes, such as transcription and chromatin remodeling. Implementation of regulatory elements that link DNA methylation to gene expression and other epigenetic modifications.
Dynamic Adjustment Codes: Creation of codes that allow for dynamic adjustments in DNA methylation patterns during cell differentiation or in response to developmental cues. Development of regulatory elements that enable the reprogramming of DNA methylation in specific genomic regions.
Feedback Control Languages: Generation of languages that enable DNMTs and associated proteins to communicate feedback signals regarding methylation levels and patterns. Implementation of codes that regulate the activity of DNMTs based on the cellular context and methylation status.
Hierarchical Regulatory Languages: Establishment of hierarchical codes that prioritize the methylation of certain genomic regions over others, guiding the distribution of methylation patterns. These regulatory codes and languages work together to ensure the accurate establishment, maintenance, and adaptation of DNA methylation patterns.
The orchestrated interplay of these codes highlights the complexity of the regulatory networks that would need to be in place to support the functionality of DNA methylation. Proponents of intelligent design argue that the simultaneous emergence and integration of these regulatory codes points toward an intentional design rather than a gradual, stepwise evolutionary process.
How did the mechanisms of DNA methylation evolve to contribute to cellular differentiation and tissue-specific functions?
The mechanisms of DNA methylation are believed to have evolved as a crucial epigenetic regulatory system that contributes to cellular differentiation and tissue-specific functions. While the exact evolutionary steps are not fully understood, there are several proposed ways in which DNA methylation mechanisms could have evolved to play a role in shaping the complexity of multicellular organisms:
Development of Cell Identity: DNA methylation likely emerged as a way to establish and maintain distinct cell identities within multicellular organisms. As cells began to specialize for different functions, DNA methylation patterns could have been utilized to lock in specific gene expression profiles that define cell types.
Tissue-Specific Gene Expression: Over time, DNA methylation could have been refined to silence or activate specific genes in a tissue-specific manner. This would allow different tissues to have unique gene expression profiles, enabling them to carry out their specialized functions while sharing the same genomic information.
Adaptation to Environmental Changes: DNA methylation may have evolved as a mechanism to enable organisms to adapt to changing environmental conditions. By modifying DNA methylation patterns in response to external cues, organisms could fine-tune their gene expression to better suit the current environment.
Prevention of Transposable Element Activity: One proposed function of DNA methylation is to suppress the activity of transposable elements (TEs), which are mobile genetic elements that can disrupt gene function. As organisms evolved, the need to control TE activity could have driven the development of DNA methylation as a defense mechanism.
Regulation of Developmental Processes: DNA methylation likely evolved to regulate key developmental processes, such as embryogenesis and organ formation. By modulating the timing and extent of DNA methylation changes, organisms could ensure proper development and tissue morphogenesis.
Evolution of Complex Traits: As organisms evolved more complex traits and adaptations, DNA methylation could have played a role in orchestrating these changes. For example, the evolution of novel features like limb development in vertebrates could involve coordinated changes in DNA methylation to support these morphological shifts.
Cellular Memory and Epigenetic Inheritance: DNA methylation's ability to maintain stable epigenetic memory across cell divisions could have provided a means for cells to remember their lineage and developmental history. This could contribute to maintaining tissue-specific functions and identities over generations.
Emergence of Regulatory Networks: As DNA methylation mechanisms evolved, they likely became integrated with other epigenetic modifications, transcription factors, and signaling pathways. This integration could have led to the formation of complex regulatory networks that govern cellular differentiation and tissue-specific functions.
Overall, the evolution of DNA methylation as a regulatory mechanism is likely intertwined with the emergence of multicellularity and the need for organisms to efficiently control gene expression in diverse cell types. The gradual refinement and utilization of DNA methylation as part of these regulatory networks could have enabled organisms to achieve higher levels of complexity and specialization. While the exact evolutionary path remains a subject of ongoing research, the integration of DNA methylation into cellular processes is seen as a remarkable example of how epigenetic mechanisms contribute to the diversity and functionality of living organisms.
Is there scientific evidence supporting the idea that DNA Methylation was brought about by the process of evolution?
The step-by-step evolution of DNA methylation faces considerable challenges due to its intricate complexity and the interdependence of its mechanisms. DNA methylation isn't an isolated feature but a system that requires multiple components to work together. It's more reasonable to consider an all-at-once, fully operational instantiation for the origin of DNA methylation, given the following considerations:
Interdependence of Mechanisms: DNA methylation involves a network of regulatory codes, languages, and proteins that must function seamlessly. The methylation machinery, recognition proteins, chromatin modifiers, and transcription factors all need to work in harmony from the outset.
Epigenetic Information System: DNA methylation is an information-based system that regulates gene expression and cellular function. The creation of complex codes, the machinery to read and interpret them, and associated regulatory elements would be challenging to evolve gradually.
Emergence of Regulatory Pathways: DNA methylation's role in gene regulation and cellular differentiation requires intricate signaling and communication pathways. These pathways, including developmental, environmental, and stress-responsive signaling, would need to be fully functional to orchestrate proper DNA methylation patterns.
Absence of Intermediate Function: Intermediate stages in the evolution of DNA methylation might lack clear functional benefit. Methylation patterns are specific and coordinated, serving as integral parts of gene regulatory networks. Intermediate stages with partial methylation functionality wouldn't offer a selective advantage.
Evolution of Adaptive Complexity: DNA methylation's role in adaptation, developmental regulation, and genome stability suggests a high level of adaptive complexity. This complexity argues for a designed system that was purposefully instantiated from the start.
In summary, the intricate interdependence, regulatory codes, and immediate functional requirement of DNA methylation make stepwise evolution unlikely. An intelligent design perspective suggests that the various components, mechanisms, codes, and signaling pathways required for DNA methylation had to be created simultaneously to ensure its critical roles in gene expression and cellular function.
Irreducibility and Interdependence of the systems to instantiate and operate DNA Methylation
The manufacturing, signaling, and regulatory codes and languages involved in the process of creating, developing, and operating DNA methylation are profoundly interconnected and indispensable. They form a complex system that functions harmoniously to facilitate this epigenetic process. From an intelligent design perspective, the simultaneous instantiation of these components is a more reasonable explanation than their gradual evolution due to the following reasons:
Manufacturing and Recognition Codes: DNA methylation relies on accurate recognition of specific DNA sequences by DNA methyltransferases. This recognition code ensures targeted methylation and prevents random modification. Without precise recognition, methylation lacks functional specificity.
Regulatory Codes and Languages: Regulatory codes determine the timing and location of DNA methylation. These codes interact with cellular signals, developmental cues, and environmental factors. Regulatory languages involve intricate interactions with transcription factors and other epigenetic modifications. Without coordinated regulation, DNA methylation wouldn't be contextually responsive for gene regulation.
Signaling Pathways and Communication: DNA methylation is influenced by signaling pathways that convey cellular status and environmental changes. These pathways, such as developmental and stress signaling, communicate with the DNA methylation machinery. Cross-communication ensures that epigenetic changes align with the cell's overall context.
Chromatin Remodeling and Structural Codes: DNA methylation interacts with chromatin structure, affecting gene expression. The interplay between methyl groups and histone modifications impacts chromatin accessibility. These structural codes determine the functional impact of DNA methylation on gene regulation.
Epigenetic Memory and Inheritance: DNA methylation patterns are inherited through cell divisions, contributing to epigenetic memory. The machinery involved, along with recognition codes, ensures accurate pattern transmission. Without proper maintenance, DNA methylation's role in cellular memory would be compromised.
Immediate Functionality and Adaptive Complexity: The interdependence of these components underscores the immediate functional necessity of DNA methylation. Gradual evolution would involve non-functional intermediates. The simultaneous creation of these codes, languages, and mechanisms is more plausible, as it would confer the gene regulatory precision required for proper cellular function.
In summary, the intricate interplay among manufacturing, signaling, and regulatory elements in DNA methylation argues for a fully operational system from the start. The complexity and the absence of functional intermediates suggest that these components were intentionally created together, aligning with the concept of an intelligently designed origin.
Once is instantiated and operational, what other intra and extracellular systems is DNA Methylation interdependent with?
Once DNA methylation is instantiated and operational, it becomes interdependent with several other intra and extracellular systems, reflecting its central role in gene regulation and cellular function.
Intracellular Systems
Chromatin Remodeling Complexes: DNA methylation interacts with chromatin remodeling complexes to regulate gene expression. The interplay between DNA methylation and histone modifications influences chromatin structure and accessibility.
Transcription Factors: Transcription factors recognize specific DNA sequences and interact with DNA methylation patterns. This interaction affects the binding and activity of transcription factors, ultimately influencing gene expression levels.
Histone Modifications: DNA methylation and histone modifications collaborate to regulate gene expression. These epigenetic marks work together to establish a complex regulatory landscape.
DNA Repair Machinery: DNA methylation can impact DNA repair processes. The DNA repair machinery responds to DNA damage and mutations, and the presence of DNA methylation can influence repair efficiency.
Extracellular Systems
Cell-Cell Communication: DNA methylation patterns can be influenced by extracellular signals, such as growth factors and cytokines. These signals can trigger changes in gene expression patterns through modifications in DNA methylation.
Epigenetic Inheritance: DNA methylation patterns can be inherited across generations and play a role in epigenetic inheritance. These patterns can impact the developmental trajectory of offspring.
Environmental Adaptation
DNA methylation can respond to environmental cues and stressors, leading to changes in gene expression patterns. This adaptability helps organisms respond to changing conditions.
Developmental Processes
Cell Differentiation: DNA methylation is crucial for cell differentiation, allowing cells with identical genetic material to take on distinct roles and functions in tissues and organs.
Embryonic Development: DNA methylation plays a role in embryonic development by guiding cell fate determination and tissue formation.
Tissue-Specific Gene Expression: DNA methylation contributes to tissue-specific gene expression patterns, enabling cells to specialize and function in specific tissue contexts.
Epigenetic Stability
Epigenetic Memory: DNA methylation can serve as a form of epigenetic memory, preserving gene expression patterns over multiple cell divisions and generations.
Genome Stability
DNA methylation can help maintain genome stability by silencing transposable elements and preventing their harmful effects on the genome.
These interdependencies highlight the integrated nature of DNA methylation within cellular processes, development, and adaptation to the environment. The complexity of these relationships underscores the challenges that an incremental stepwise evolution of DNA methylation would face, suggesting a more plausible scenario of simultaneous instantiation to ensure functional coherence and precision from the outset.
1. DNA methylation is interdependent with chromatin remodeling complexes, transcription factors, histone modifications, and DNA repair machinery. These systems collectively regulate gene expression and genome stability.
2. DNA methylation responds to extracellular signals and influences epigenetic inheritance, adaptation to the environment, and developmental processes like cell differentiation and tissue-specific gene expression.
3. The complexity of these interdependencies and their immediate functional requirement implies a fully operational system from the outset.
Conclusion: In light of this, a plausible explanation is that DNA methylation, with its regulatory codes, signaling networks, and cross-system communication, was deliberately designed as an integrated framework to ensure precise gene regulation, cellular differentiation, and environmental responsiveness. The intricate interlocking of these systems, each relying on the others for meaningful function, is more consistent with the concept of an intelligently orchestrated origin rather than a stepwise evolutionary process.
DNA methylation is an epigenetic modification that involves the addition of a methyl group to the cytosine base of a DNA molecule, typically occurring at cytosine-guanine dinucleotide sequences (CpG sites). This modification is catalyzed by enzymes called DNA methyltransferases. DNA methylation plays a crucial role in regulating gene expression by modulating the accessibility of DNA to transcription factors and other regulatory proteins. In DNA methylation, a methyl group (CH3) is added to the carbon-5 position of the cytosine ring. This modification can lead to changes in chromatin structure, making the associated DNA regions more condensed and less accessible for transcriptional machinery. As a result, genes in methylated regions are often silenced or have reduced transcriptional activity. Conversely, unmethylated regions are generally associated with active gene expression.
Importance in Biological Systems
DNA methylation is essential for various biological processes and cellular functions:
Gene Regulation: DNA methylation plays a central role in controlling gene expression. Methylated DNA segments prevent the binding of transcription factors and other regulatory proteins, inhibiting gene transcription.
Cellular Identity and Differentiation: DNA methylation patterns are established during development and are crucial for determining cell identity. They contribute to the differentiation of cells into various specialized types in multicellular organisms.
X-Chromosome Inactivation: In females, DNA methylation is involved in X-chromosome inactivation, a process that compensates for the presence of two X chromosomes. One X chromosome in each cell becomes highly methylated and largely inactive.
Epigenetic Memory: DNA methylation patterns can be heritably passed on to daughter cells during cell division. This epigenetic memory contributes to maintaining cell identity and lineage-specific gene expression.
Tumor Suppression: DNA methylation can act as a tumor suppressor mechanism by silencing genes involved in cell cycle regulation and DNA repair. Aberrant DNA methylation is associated with cancer development.
Developmental Processes Shaping Organismal Form and Function
DNA methylation is particularly important in shaping organismal form and function during developmental processes:
Embryonic Development: DNA methylation patterns undergo dynamic changes during embryonic development. They guide the differentiation of cells into distinct lineages, leading to the formation of tissues and organs.
Tissue-Specific Gene Expression: DNA methylation contributes to establishing tissue-specific gene expression profiles. Different cell types exhibit unique methylation patterns that help maintain their specialized functions.
Imprint Control: DNA methylation controls imprinted genes, which are expressed in a parent-of-origin-specific manner. Imprinting influences aspects of growth, development, and behavior.
Adaptation to Environment: DNA methylation can respond to environmental cues and influence gene expression accordingly. This process allows organisms to adapt to changing environmental conditions.
DNA methylation is a fundamental epigenetic modification that plays a pivotal role in gene regulation, cellular differentiation, developmental processes, and the establishment of cell identity. Its dynamic and context-dependent nature makes it a critical factor in shaping the form and function of organisms throughout their life cycles.
What is the significance of DNA methylation in regulating gene expression and cell differentiation during development?
DNA methylation plays a significant role in regulating gene expression and cell differentiation during development by influencing the accessibility of genes to the transcriptional machinery and guiding cells toward specific lineages. Here's how DNA methylation contributes to these processes:
Gene Expression Regulation
Silencing Gene Expression: DNA methylation at promoter regions of genes can physically block the binding of transcription factors and other regulatory proteins. This prevents the initiation of transcription, effectively silencing gene expression.
Stable Repression: DNA methylation provides a stable and heritable form of gene repression. Once established, methylation patterns can be maintained through DNA replication and cell division, ensuring long-term control over gene expression.
Tissue-Specific Expression: DNA methylation patterns are often tissue-specific. By methylating certain genes in specific cell types, the expression of those genes can be restricted to certain tissues, contributing to the development of distinct cell lineages.
Cell Differentiation and Development
Epigenetic Memory: DNA methylation patterns acquired during early development can serve as epigenetic memory, ensuring that cells maintain their specialized functions as they divide and differentiate. This contributes to the stability of cell identities.
Lineage Commitment: DNA methylation patterns guide cells toward specific lineages during differentiation. By silencing genes associated with alternative cell fates, DNA methylation helps commit cells to their intended developmental pathway.
Imprinting and Parental Alleles: DNA methylation is involved in genomic imprinting, where certain genes are expressed based on their parental origin. Imprinting is critical for proper development, as it ensures the appropriate expression of genes that influence growth and metabolism.
X-Chromosome Inactivation: DNA methylation is responsible for X-chromosome inactivation in females. It ensures that one of the two X chromosomes in each cell is largely silenced, preventing an imbalance in gene dosage between males and females.
Cell Identity Maintenance: DNA methylation helps maintain cell identity by ensuring that only relevant genes are expressed in a given cell type. Changes in DNA methylation patterns can disrupt this identity and lead to aberrant cell behavior.
Adaptation and Plasticity
Environmental Response: DNA methylation can be influenced by environmental factors, allowing organisms to adapt to changing conditions. Changes in methylation patterns can modulate gene expression in response to environmental cues.
Developmental Plasticity: During early development, DNA methylation can confer a degree of plasticity, allowing cells to adopt different fates under specific conditions. As development progresses, methylation patterns become more stable and guide cellular differentiation.
How are DNA methylation patterns established and maintained through cell divisions?
The establishment and maintenance of DNA methylation patterns through cell divisions involve a combination of enzymatic processes, DNA replication, and epigenetic memory mechanisms. These processes ensure the faithful transmission of epigenetic information from one generation of cells to the next. Here's how DNA methylation patterns are established and preserved:
Establishment of DNA Methylation Patterns
De Novo Methylation: During early embryonic development, de novo DNA methylation occurs, where specific regions of the genome are marked with methyl groups. This process is catalyzed by enzymes called DNA methyltransferases (DNMTs).
Maintenance Methylation: Maintenance methylation refers to the replication-dependent addition of methyl groups to the newly synthesized DNA strand during DNA replication. The enzyme DNMT1 plays a crucial role in this process. DNMT1 recognizes hemi-methylated CpG sites (one methylated strand and one unmethylated strand) and adds a methyl group to the newly synthesized strand, thereby maintaining the methylation pattern after DNA replication.
Maintenance of DNA Methylation Patterns Through Cell Divisions
Epigenetic Memory: DNA methylation patterns can be maintained across multiple cell divisions due to epigenetic memory mechanisms. These mechanisms involve interactions between DNMTs and histone modifications that help stabilize the methylation pattern.
DNMT1 and Replication: As mentioned earlier, DNMT1 plays a key role in maintaining DNA methylation patterns during cell division. It ensures that the newly synthesized DNA strand is methylated based on the pattern of the template strand. This process helps preserve the original methylation pattern in daughter cells.
Histone Modifications: Histone modifications and DNA methylation are closely linked. Certain histone modifications contribute to the recruitment of DNMTs to specific genomic regions. This coordination helps ensure that DNA methylation is faithfully maintained during replication and cell divisions.
Passive Demethylation Prevention: Passive demethylation occurs when methylated cytosines are not maintained during DNA replication. Histone modifications and DNMT1 activity prevent passive demethylation by ensuring that newly synthesized DNA strands are promptly methylated.
Epigenetic Plasticity and Adaptation
Environmental Influence: While DNA methylation patterns are generally stable, certain environmental factors can influence them. Environmental cues can lead to changes in DNA methylation patterns, allowing cells to adapt to new conditions and influences.
Epigenetic Variability: Despite the stability of DNA methylation, some degree of epigenetic variability exists between individuals and cell types. This variability can contribute to phenotypic diversity and may have implications for health and disease.
DNA methylation patterns are established during early development through de novo methylation and are then maintained through cell divisions via processes like maintenance methylation. Epigenetic memory mechanisms, interactions between DNMTs and histone modifications, and the faithful action of DNMT1 play crucial roles in preserving the methylation pattern. This stability contributes to the maintenance of cell identity, gene expression patterns, and proper development throughout the life of an organism.

DNA methylation plays a pivotal role in the epigenetic regulation of gene expression. Specifically, the addition of a methyl group to the carbon 5 position of cytosine within cytosine-phosphate-guanine (CpG) and other nucleotide sequences has profound effects on gene activity. This epigenetic modification acts as a molecular "off" switch, inhibiting the binding of transcription factors to gene promoters. This simple chemical alteration introduces a layer of complexity to the genome's regulatory landscape, influencing how genes are read and interpreted by the cellular machinery. By methylating CpG sites in promoter regions, DNA methylation interferes with the recruitment of transcription factors—proteins that play a key role in initiating gene transcription. The binding of transcription factors to promoter regions is a crucial step in the process of gene activation. DNA methylation at these sites essentially creates a physical barrier that hinders the transcription factors from binding effectively, leading to reduced or silenced gene expression. This mechanism of DNA methylation-induced gene silencing has far-reaching implications for cellular processes, development, and health. It enables cells to fine-tune gene expression patterns, allowing them to respond to environmental cues, differentiate into specialized cell types, and maintain epigenetic memory. The intricate orchestration of DNA methylation and its interactions with other epigenetic marks, chromatin structure, and transcriptional machinery illustrate the elegant complexity of gene regulation within the cell. This interplay underscores the sophisticated control mechanisms that exist to ensure the appropriate functioning of our genetic material.
Appearance of DNA Methylation in the evolutionary timeline
The appearance of DNA methylation in the evolutionary timeline is a complex topic that involves speculation and ongoing research. While the exact timing and sequence of events are not fully understood, here is a general overview of the hypothesized appearance of DNA methylation in the evolutionary timeline:
Early Life Forms: DNA methylation is not likely to have played a significant role in early cellular life forms, as they lacked the organized cellular structures and regulatory mechanisms present in modern organisms.
Emergence of Prokaryotes: DNA methylation is hypothesized to have emerged relatively late in the evolution of life. Initial forms of DNA methylation would have been simple and limited in scope.
In prokaryotes, DNA methylation could have potentially played a role in the regulation of gene expression, although the extent and functions are not well established.
Transition to Eukaryotes: The appearance of eukaryotic cells would have brought about more complex regulatory mechanisms, including epigenetic modifications like DNA methylation. DNA methylation would have gained importance in eukaryotes as a mechanism for gene regulation, controlling processes such as cell differentiation and development.
Evolution of Multicellularity: With the emergence of multicellular organisms, DNA methylation would have become more intricate and diverse in its functions. The regulation of cell differentiation, tissue-specific gene expression, and the establishment of cell identities would have become crucial, making DNA methylation a key player in these processes.
Vertebrate Evolution and Complexity: Vertebrates, including mammals, exhibit highly complex DNA methylation patterns associated with various cellular processes. In vertebrates, DNA methylation plays roles in genomic imprinting, X-chromosome inactivation, and the regulation of tissue-specific gene expression.
Expansion of Functions and Epigenetic Landscape: Over time, DNA methylation would have evolved to have broader functions, influencing not only gene expression but also genome stability, response to environmental cues, and adaptation. The epigenetic landscape would have become more complex as additional enzymes and regulatory mechanisms evolved to modulate DNA methylation patterns.
It's important to note that the timeline presented here is speculative, and the actual appearance and evolution of DNA methylation may have occurred differently. The study of DNA methylation's evolutionary history is an active area of research, and ongoing discoveries continue to shape our understanding of its origin and function across different species.
De Novo Genetic Information necessary to instantiate DNA Methylation
Creating the mechanisms of DNA methylation de novo involves the generation and integration of new genetic information to establish the enzymes, regulatory elements, and recognition systems required for DNA methylation. Here's a simplified overview of the hypothetical process:
Generation of Enzymes: New genetic information would need to code for enzymes known as DNA methyltransferases (DNMTs). These enzymes recognize specific DNA sequences and catalyze the addition of methyl groups to cytosine bases.
Regulatory Elements and Promoters: Regulatory elements, including promoters and enhancers, would need to emerge to control the expression of DNMT genes. These elements ensure that DNMTs are produced at the right time and in the right cells.
Target Recognition Sequences: New genetic information would need to provide the sequences that DNMTs recognize as their target sites for methylation (CpG sites). These sequences would need to be distributed appropriately throughout the genome.
Accessory Proteins: The genetic information would also need to encode accessory proteins that interact with DNMTs, ensuring proper enzymatic activity, binding, and recruitment to target sites.
Epigenetic Memory Mechanisms: To maintain DNA methylation patterns during cell divisions, epigenetic memory mechanisms would need to emerge. This could involve interactions between DNA methylation and histone modifications or the development of specialized proteins that facilitate maintenance.
Establishing Methylation Patterns: The genetic information would need to specify the initial methylation patterns, indicating which regions of the genome should be methylated and which should remain unmethylated.
Integration of Regulatory Networks: New genetic information would need to establish regulatory networks that link DNA methylation to other cellular processes, such as transcription, DNA repair, and chromatin remodeling.
Integration with Replication Machinery: The genetic information would need to coordinate with the DNA replication machinery to ensure that newly synthesized DNA strands are methylated in a manner consistent with the original pattern.
Environmental Sensitivity: If the hypothetical system is designed to respond to environmental cues, additional genetic information would be needed to link DNA methylation patterns to specific environmental signals.
Integration into the Genome: The new genetic information would need to integrate seamlessly into the existing genome to ensure that the mechanisms of DNA methylation function harmoniously with other cellular processes.
This hypothetical process involves the orchestrated creation of genes, regulatory elements, sequences, and mechanisms that work together to establish and maintain DNA methylation.
It's important to note that this description simplifies a complex process and doesn't account for the intricate interactions and regulatory networks that would be required for the functional instantiation of DNA methylation.
Manufacturing codes and languages that would have to emerge and be employed to instantiate DNA Methylation
To transition from an organism without DNA methylation to one with a fully developed DNA methylation system, a complex set of manufacturing codes and languages would need to be created, instantiated, and orchestrated. These non-genetic components are integral for the establishment, regulation, and maintenance of the DNA methylation machinery. Here's an explanation of the manufacturing codes and languages involved in this process:
Enzymatic Codes and Protein Assembly: Generation of codes that dictate the precise sequence and structure of DNA methyltransferase enzymes (DNMTs). Creation of molecular chaperones that aid in the correct folding and assembly of DNMTs, ensuring their functional three-dimensional structure.
Regulatory Elements and Promoter Codes: Establishment of codes that define the regulatory elements controlling the expression of genes encoding DNMTs. Development of promoter codes that determine the strength, timing, and tissue-specificity of DNMT gene expression.
CpG Recognition Codes: Creation of codes that specify the recognition sequences for DNMTs to identify and methylate CpG sites in DNA. Design of binding motifs within DNMTs that enable accurate recognition and binding to CpG-rich regions.
Epigenetic Inheritance Codes: Generation of codes that enable the transmission of DNA methylation patterns from parent to daughter cells during DNA replication. Implementation of feedback loops and stability codes to prevent random changes in methylation patterns.
Interplay and Integration Codes: Development of codes that facilitate crosstalk between DNA methylation and other epigenetic modifications, such as histone modifications. Creation of codes that enable coordinated regulation of gene expression through the interplay of DNA methylation and chromatin structure.
Environmental Responsiveness Mechanisms (Optional): Design of codes that allow the DNA methylation system to respond to environmental cues, leading to dynamic changes in methylation patterns. Implementation of decoding mechanisms that interpret external signals and trigger appropriate adjustments in DNA methylation.
Feedback Control Languages: Establishment of languages that enable DNMTs and associated proteins to communicate feedback signals regarding methylation levels and patterns. Development of codes that regulate the activity of DNMTs based on the cellular context and methylation status.
Integration and Organization Codes: Creation of codes that ensure the proper integration of the DNA methylation machinery into the cellular context. Design of organizational codes that guide the spatial arrangement and functional interactions of DNMTs and related components. The orchestrated creation and integration of these manufacturing codes and languages are essential to establish a functional DNA methylation system.
This intricate interplay among diverse components highlights the complexity of the process and raises questions about the simultaneous emergence and coordination of these codes. Proponents of intelligent design argue that the presence of such sophisticated and interdependent systems supports the notion of purposeful design rather than a gradual, step-by-step evolutionary process.
Epigenetic Regulatory Mechanisms necessary to be instantiated to create DNA Methylation
The development of DNA methylation from scratch would require the creation and subsequent utilization of intricate epigenetic regulatory systems. These systems would work in tandem to establish, regulate, and maintain DNA methylation. Here's an overview of the epigenetic regulation and the collaborative systems involved:
Epigenetic Regulatory Systems
DNA Methyltransferases (DNMTs) and Regulatory Elements: Creation of regulatory elements that control the expression of genes encoding DNMTs. Design of mechanisms that ensure DNMT expression is fine-tuned based on developmental stages and cellular contexts.
Methylation Target Recognition and Binding: Generation of mechanisms that enable DNMTs to recognize specific DNA sequences (CpG sites) for methylation. Establishment of codes that ensure accurate binding and proper positioning of DNMTs at target sites.
Epigenetic Memory and Maintenance: Design of epigenetic memory systems that retain DNA methylation patterns during cell divisions. Integration of mechanisms that coordinate DNA methylation maintenance with DNA replication and chromatin remodeling.
Integration with Chromatin Modifications: Creation of codes that allow crosstalk and coordination between DNA methylation and histone modifications. Development of molecular mechanisms that modulate chromatin structure and accessibility in response to DNA methylation changes.
Collaborative Systems
Chromatin Remodeling Complexes: Collaborative interaction with chromatin remodeling complexes to expose target regions for methylation and facilitate proper DNMT binding.
Histone Modification Systems: Joint operation with histone modification systems to create an epigenetic landscape that guides DNA methylation to specific genomic regions.
DNA Repair and Replication Machinery: Collaborative coordination with DNA repair and replication machinery to ensure accurate maintenance of DNA methylation patterns during cell divisions.
Transcriptional Regulatory Networks: Collaboration with transcriptional regulatory networks to influence gene expression patterns through the modulation of DNA methylation status.
Environmental Sensing and Adaptation (Optional): Joint functioning with environmental sensing mechanisms to allow for dynamic changes in DNA methylation patterns in response to external cues.
Cell Signaling Pathways: Collaboration with cell signaling pathways to integrate cellular signals that influence DNA methylation patterns based on physiological conditions.
Epigenetic Enzyme Networks: Collaborative interaction with other epigenetic enzymes, such as histone modifiers and chromatin remodelers, to orchestrate coordinated changes in chromatin structure.
The successful establishment of DNA methylation would require an intricate interplay among these epigenetic regulatory systems and collaborative mechanisms. Each component must be precisely created and integrated to ensure accurate DNA methylation patterns, stable epigenetic memory, and proper functioning within the cellular context. The complexity of these interdependent systems raises questions about their simultaneous emergence and coordinated operation, which is evidence for the purposeful and coordinated origin of such complex cellular processes.
Signaling Pathways necessary to create, and maintain DNA Methylation
The emergence of DNA methylation from scratch would necessitate the creation and subsequent involvement of signaling pathways that orchestrate and regulate this epigenetic modification. These signaling pathways would be interconnected, interdependent, and engage in crosstalk with each other and other biological systems to ensure accurate DNA methylation patterns. Here's an overview of the hypothetical signaling pathways and their interactions:
Developmental Signaling Pathways
Creation of signaling pathways that respond to developmental cues and guide the establishment of DNA methylation patterns in specific cells and tissues. Interdependence with transcription factors and chromatin modifiers to coordinate gene expression and epigenetic regulation.
Environmental Sensing Pathways: Generation of pathways that sense environmental cues and enable dynamic changes in DNA methylation patterns in response to external conditions. Interconnection with cellular stress responses and adaptive mechanisms to optimize cellular function.
DNA Damage and Repair Signaling: Establishment of signaling pathways that detect DNA damage and trigger DNA repair mechanisms. Crosstalk with DNA methylation maintenance systems to ensure accurate restoration of methylation patterns after repair.
Cell Signaling Cascades: Creation of cascades that transduce extracellular signals, such as growth factors or hormones, into intracellular responses that influence DNA methylation. Interactions with transcriptional regulators and chromatin modifiers to modulate gene expression and epigenetic states.
Epigenetic Cross-Talk Pathways: Generation of pathways that facilitate communication between different epigenetic modifications, such as DNA methylation and histone modifications. Interplay with chromatin remodeling complexes to establish coordinated changes in chromatin structure.
Replication and Chromatin Dynamics Pathways: Development of pathways that synchronize DNA methylation maintenance with DNA replication and chromatin dynamics. Coordination with histone modification pathways to ensure proper chromatin packaging and accessibility.
Transcriptional Feedback Loops: Establishment of feedback loops that link DNA methylation status with gene expression levels. Interdependence with transcription factors and RNA processing pathways to fine-tune gene regulation.
Cellular Stress and Homeostasis Pathways: Creation of pathways that monitor cellular stress and maintain homeostasis by adjusting DNA methylation patterns. Crosstalk with cell cycle checkpoints and metabolic pathways to ensure cellular integrity.
These signaling pathways would interact extensively with each other and with broader biological systems to ensure the accurate establishment and maintenance of DNA methylation patterns. The complexity and interconnectedness of these pathways underscore the intricate regulatory networks that would need to be simultaneously created and coordinated to support the emergence of DNA methylation. Proponents of intelligent design argue that the orchestrated integration of these pathways points toward a purposeful and intentional design rather than a stepwise evolutionary process.
Regulatory codes necessary for maintenance and operation of DNA Methylation
The maintenance and operation of DNA methylation would require the instantiation and subsequent involvement of intricate regulatory codes and languages that govern various aspects of this epigenetic modification. These codes and languages play a crucial role in ensuring the stability, accuracy, and responsiveness of DNA methylation patterns. Here's an overview of the regulatory codes and languages involved:
Maintenance and Inheritance Codes: Establishment of codes that facilitate the faithful transmission of DNA methylation patterns to daughter cells during DNA replication. Development of mechanisms that ensure the maintenance of established methylation patterns through cell divisions.
Methylation Stability Codes: Creation of codes that prevent random changes in DNA methylation patterns, ensuring stability and epigenetic memory. Implementation of feedback loops that monitor and rectify deviations from the desired methylation state.
Context-Dependent Codes: Generation of codes that allow for context-dependent DNA methylation, meaning that patterns can be adapted to specific cellular environments or developmental stages. Incorporation of regulatory elements that respond to cell type-specific cues to establish appropriate methylation patterns.
Environmental Responsiveness Codes (Optional): Design of codes that enable DNA methylation patterns to be modulated in response to external environmental signals. Integration of regulatory networks that interpret environmental cues and trigger changes in DNA methylation status.
Crosstalk and Communication Codes: Establishment of codes that facilitate communication between DNA methylation machinery and other cellular processes, such as transcription and chromatin remodeling. Implementation of regulatory elements that link DNA methylation to gene expression and other epigenetic modifications.
Dynamic Adjustment Codes: Creation of codes that allow for dynamic adjustments in DNA methylation patterns during cell differentiation or in response to developmental cues. Development of regulatory elements that enable the reprogramming of DNA methylation in specific genomic regions.
Feedback Control Languages: Generation of languages that enable DNMTs and associated proteins to communicate feedback signals regarding methylation levels and patterns. Implementation of codes that regulate the activity of DNMTs based on the cellular context and methylation status.
Hierarchical Regulatory Languages: Establishment of hierarchical codes that prioritize the methylation of certain genomic regions over others, guiding the distribution of methylation patterns. These regulatory codes and languages work together to ensure the accurate establishment, maintenance, and adaptation of DNA methylation patterns.
The orchestrated interplay of these codes highlights the complexity of the regulatory networks that would need to be in place to support the functionality of DNA methylation. Proponents of intelligent design argue that the simultaneous emergence and integration of these regulatory codes points toward an intentional design rather than a gradual, stepwise evolutionary process.
How did the mechanisms of DNA methylation evolve to contribute to cellular differentiation and tissue-specific functions?
The mechanisms of DNA methylation are believed to have evolved as a crucial epigenetic regulatory system that contributes to cellular differentiation and tissue-specific functions. While the exact evolutionary steps are not fully understood, there are several proposed ways in which DNA methylation mechanisms could have evolved to play a role in shaping the complexity of multicellular organisms:
Development of Cell Identity: DNA methylation likely emerged as a way to establish and maintain distinct cell identities within multicellular organisms. As cells began to specialize for different functions, DNA methylation patterns could have been utilized to lock in specific gene expression profiles that define cell types.
Tissue-Specific Gene Expression: Over time, DNA methylation could have been refined to silence or activate specific genes in a tissue-specific manner. This would allow different tissues to have unique gene expression profiles, enabling them to carry out their specialized functions while sharing the same genomic information.
Adaptation to Environmental Changes: DNA methylation may have evolved as a mechanism to enable organisms to adapt to changing environmental conditions. By modifying DNA methylation patterns in response to external cues, organisms could fine-tune their gene expression to better suit the current environment.
Prevention of Transposable Element Activity: One proposed function of DNA methylation is to suppress the activity of transposable elements (TEs), which are mobile genetic elements that can disrupt gene function. As organisms evolved, the need to control TE activity could have driven the development of DNA methylation as a defense mechanism.
Regulation of Developmental Processes: DNA methylation likely evolved to regulate key developmental processes, such as embryogenesis and organ formation. By modulating the timing and extent of DNA methylation changes, organisms could ensure proper development and tissue morphogenesis.
Evolution of Complex Traits: As organisms evolved more complex traits and adaptations, DNA methylation could have played a role in orchestrating these changes. For example, the evolution of novel features like limb development in vertebrates could involve coordinated changes in DNA methylation to support these morphological shifts.
Cellular Memory and Epigenetic Inheritance: DNA methylation's ability to maintain stable epigenetic memory across cell divisions could have provided a means for cells to remember their lineage and developmental history. This could contribute to maintaining tissue-specific functions and identities over generations.
Emergence of Regulatory Networks: As DNA methylation mechanisms evolved, they likely became integrated with other epigenetic modifications, transcription factors, and signaling pathways. This integration could have led to the formation of complex regulatory networks that govern cellular differentiation and tissue-specific functions.
Overall, the evolution of DNA methylation as a regulatory mechanism is likely intertwined with the emergence of multicellularity and the need for organisms to efficiently control gene expression in diverse cell types. The gradual refinement and utilization of DNA methylation as part of these regulatory networks could have enabled organisms to achieve higher levels of complexity and specialization. While the exact evolutionary path remains a subject of ongoing research, the integration of DNA methylation into cellular processes is seen as a remarkable example of how epigenetic mechanisms contribute to the diversity and functionality of living organisms.
Is there scientific evidence supporting the idea that DNA Methylation was brought about by the process of evolution?
The step-by-step evolution of DNA methylation faces considerable challenges due to its intricate complexity and the interdependence of its mechanisms. DNA methylation isn't an isolated feature but a system that requires multiple components to work together. It's more reasonable to consider an all-at-once, fully operational instantiation for the origin of DNA methylation, given the following considerations:
Interdependence of Mechanisms: DNA methylation involves a network of regulatory codes, languages, and proteins that must function seamlessly. The methylation machinery, recognition proteins, chromatin modifiers, and transcription factors all need to work in harmony from the outset.
Epigenetic Information System: DNA methylation is an information-based system that regulates gene expression and cellular function. The creation of complex codes, the machinery to read and interpret them, and associated regulatory elements would be challenging to evolve gradually.
Emergence of Regulatory Pathways: DNA methylation's role in gene regulation and cellular differentiation requires intricate signaling and communication pathways. These pathways, including developmental, environmental, and stress-responsive signaling, would need to be fully functional to orchestrate proper DNA methylation patterns.
Absence of Intermediate Function: Intermediate stages in the evolution of DNA methylation might lack clear functional benefit. Methylation patterns are specific and coordinated, serving as integral parts of gene regulatory networks. Intermediate stages with partial methylation functionality wouldn't offer a selective advantage.
Evolution of Adaptive Complexity: DNA methylation's role in adaptation, developmental regulation, and genome stability suggests a high level of adaptive complexity. This complexity argues for a designed system that was purposefully instantiated from the start.
In summary, the intricate interdependence, regulatory codes, and immediate functional requirement of DNA methylation make stepwise evolution unlikely. An intelligent design perspective suggests that the various components, mechanisms, codes, and signaling pathways required for DNA methylation had to be created simultaneously to ensure its critical roles in gene expression and cellular function.
Irreducibility and Interdependence of the systems to instantiate and operate DNA Methylation
The manufacturing, signaling, and regulatory codes and languages involved in the process of creating, developing, and operating DNA methylation are profoundly interconnected and indispensable. They form a complex system that functions harmoniously to facilitate this epigenetic process. From an intelligent design perspective, the simultaneous instantiation of these components is a more reasonable explanation than their gradual evolution due to the following reasons:
Manufacturing and Recognition Codes: DNA methylation relies on accurate recognition of specific DNA sequences by DNA methyltransferases. This recognition code ensures targeted methylation and prevents random modification. Without precise recognition, methylation lacks functional specificity.
Regulatory Codes and Languages: Regulatory codes determine the timing and location of DNA methylation. These codes interact with cellular signals, developmental cues, and environmental factors. Regulatory languages involve intricate interactions with transcription factors and other epigenetic modifications. Without coordinated regulation, DNA methylation wouldn't be contextually responsive for gene regulation.
Signaling Pathways and Communication: DNA methylation is influenced by signaling pathways that convey cellular status and environmental changes. These pathways, such as developmental and stress signaling, communicate with the DNA methylation machinery. Cross-communication ensures that epigenetic changes align with the cell's overall context.
Chromatin Remodeling and Structural Codes: DNA methylation interacts with chromatin structure, affecting gene expression. The interplay between methyl groups and histone modifications impacts chromatin accessibility. These structural codes determine the functional impact of DNA methylation on gene regulation.
Epigenetic Memory and Inheritance: DNA methylation patterns are inherited through cell divisions, contributing to epigenetic memory. The machinery involved, along with recognition codes, ensures accurate pattern transmission. Without proper maintenance, DNA methylation's role in cellular memory would be compromised.
Immediate Functionality and Adaptive Complexity: The interdependence of these components underscores the immediate functional necessity of DNA methylation. Gradual evolution would involve non-functional intermediates. The simultaneous creation of these codes, languages, and mechanisms is more plausible, as it would confer the gene regulatory precision required for proper cellular function.
In summary, the intricate interplay among manufacturing, signaling, and regulatory elements in DNA methylation argues for a fully operational system from the start. The complexity and the absence of functional intermediates suggest that these components were intentionally created together, aligning with the concept of an intelligently designed origin.
Once is instantiated and operational, what other intra and extracellular systems is DNA Methylation interdependent with?
Once DNA methylation is instantiated and operational, it becomes interdependent with several other intra and extracellular systems, reflecting its central role in gene regulation and cellular function.
Intracellular Systems
Chromatin Remodeling Complexes: DNA methylation interacts with chromatin remodeling complexes to regulate gene expression. The interplay between DNA methylation and histone modifications influences chromatin structure and accessibility.
Transcription Factors: Transcription factors recognize specific DNA sequences and interact with DNA methylation patterns. This interaction affects the binding and activity of transcription factors, ultimately influencing gene expression levels.
Histone Modifications: DNA methylation and histone modifications collaborate to regulate gene expression. These epigenetic marks work together to establish a complex regulatory landscape.
DNA Repair Machinery: DNA methylation can impact DNA repair processes. The DNA repair machinery responds to DNA damage and mutations, and the presence of DNA methylation can influence repair efficiency.
Extracellular Systems
Cell-Cell Communication: DNA methylation patterns can be influenced by extracellular signals, such as growth factors and cytokines. These signals can trigger changes in gene expression patterns through modifications in DNA methylation.
Epigenetic Inheritance: DNA methylation patterns can be inherited across generations and play a role in epigenetic inheritance. These patterns can impact the developmental trajectory of offspring.
Environmental Adaptation
DNA methylation can respond to environmental cues and stressors, leading to changes in gene expression patterns. This adaptability helps organisms respond to changing conditions.
Developmental Processes
Cell Differentiation: DNA methylation is crucial for cell differentiation, allowing cells with identical genetic material to take on distinct roles and functions in tissues and organs.
Embryonic Development: DNA methylation plays a role in embryonic development by guiding cell fate determination and tissue formation.
Tissue-Specific Gene Expression: DNA methylation contributes to tissue-specific gene expression patterns, enabling cells to specialize and function in specific tissue contexts.
Epigenetic Stability
Epigenetic Memory: DNA methylation can serve as a form of epigenetic memory, preserving gene expression patterns over multiple cell divisions and generations.
Genome Stability
DNA methylation can help maintain genome stability by silencing transposable elements and preventing their harmful effects on the genome.
These interdependencies highlight the integrated nature of DNA methylation within cellular processes, development, and adaptation to the environment. The complexity of these relationships underscores the challenges that an incremental stepwise evolution of DNA methylation would face, suggesting a more plausible scenario of simultaneous instantiation to ensure functional coherence and precision from the outset.
1. DNA methylation is interdependent with chromatin remodeling complexes, transcription factors, histone modifications, and DNA repair machinery. These systems collectively regulate gene expression and genome stability.
2. DNA methylation responds to extracellular signals and influences epigenetic inheritance, adaptation to the environment, and developmental processes like cell differentiation and tissue-specific gene expression.
3. The complexity of these interdependencies and their immediate functional requirement implies a fully operational system from the outset.
Conclusion: In light of this, a plausible explanation is that DNA methylation, with its regulatory codes, signaling networks, and cross-system communication, was deliberately designed as an integrated framework to ensure precise gene regulation, cellular differentiation, and environmental responsiveness. The intricate interlocking of these systems, each relying on the others for meaningful function, is more consistent with the concept of an intelligently orchestrated origin rather than a stepwise evolutionary process.


