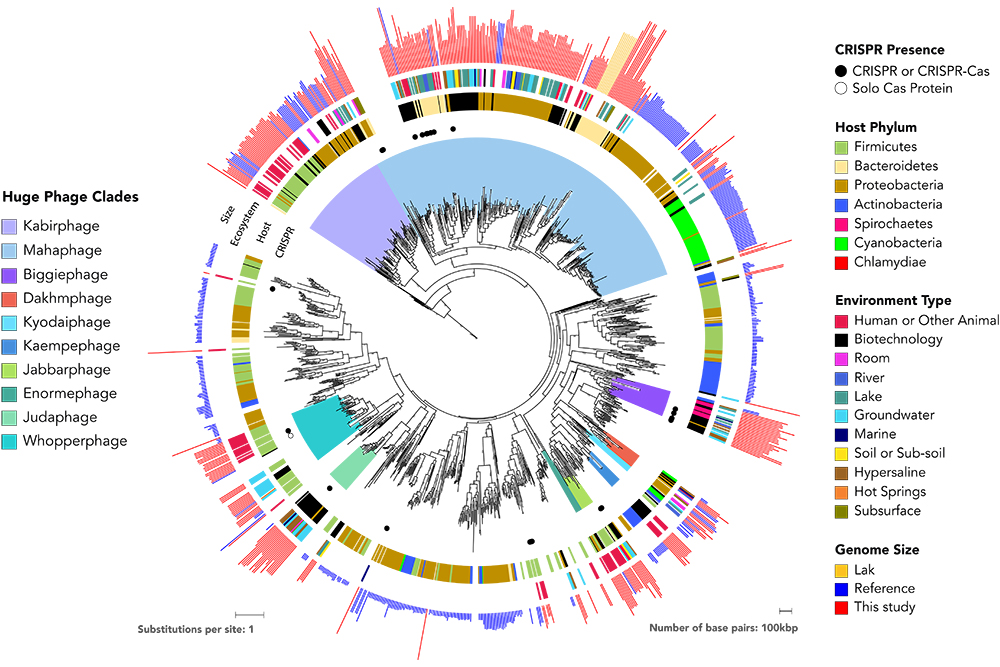
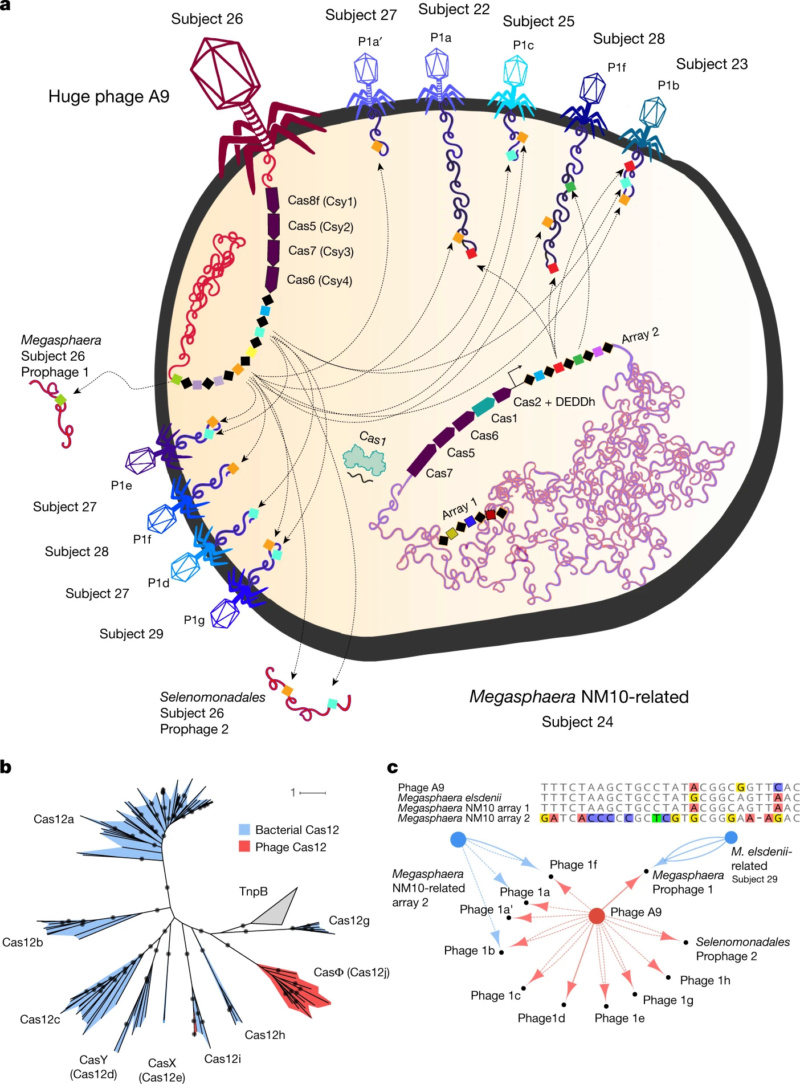
Giant Viruses Called 'Jumbo Phages'
https://reasonandscience.catsboard.com/t3256-giant-viruses-called-jumbo-phages
TESSA KOUMOUNDOUROS (2022): As antibiotic-resistant bacteria become increasingly challenging to defeat, consulting those who've been in battle with our ancient foe long before us just makes a whole lot of sense. Viruses called bacteriophages have been clashing with bacteria since long before we even existed. So researchers from the University of California, San Diego, took a closer look at some of the lesser studied jumbo phages, specifically 201phi2-1, which infects Pseudomonas chlororaphis bacteria.
Named for their large genomes of over 200,000 base pairs long (most phages that infect Pseudomonas have genomes less than 100,000 base pairs), jumbo phages have an arsenal of techniques to counter bacterial defense mechanisms. Previously research found growing a shield around its genetic material was one of these tactics, making these viruses' life cycle unique.
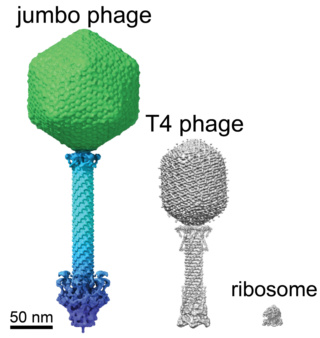
The jumbo phage compared to T4 phage virus.
Separation of genomic material from the rest of the cell contents by encasing it "was previously thought to have evolved only once in the history of life", molecular biologist Thomas Laughlin and colleagues write in their new paper. But it turns out these tiny giants beat us to it.
"It's a different kind of compartment – unlike anything we have ever seen in nature," says biophysicist Elizabeth Villa.
Usually, phages inject their genetic material into the bacteria where it floats freely in the cell's inner goo (cytoplasm) as the virus hijacks the bacteria's equipment to replicate itself. But these jumbo phages construct a separating compartment around their DNA soon after it enters its host, a bit like how our cells have a nucleus to protect our DNA.
This physically prevents the bacteria's CRISPR 'immune system' and other defensive enzymes from messing with the viral DNA. Laughlin and their team used cryo-electron microscopy and tomography to examine this compartment down to the atomic scale. The protective casing was constructed from just a single type of protein, which the researchers called chimallin, after an ancient Aztec shield.
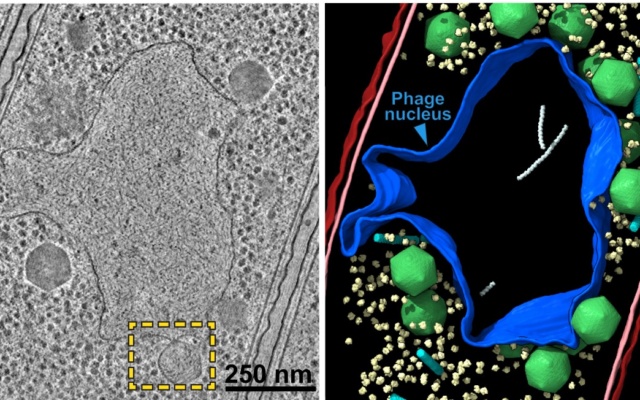
Micrograph of jumbo phage-infected bacterial cell (left) and nucleus-like compartment outlined in blue (right).
With the help of computer modeling, the researchers found the phage nucleus selectively allows molecules to pass through tiny pores – again similar to how cells control the environment around our genetic material, making this a remarkable example of convergent evolution – when completely unrelated organisms end up with a similar solution to the same biological problem.
Comment:
Convergence, another problem for evolution
https://reasonandscience.catsboard.com/t2014-convergence-another-problem-for-evolution
Biologists are uncovering numerous examples of organisms that cluster together morphologically (structurally), and yet are genetically distinct. Frogs, lizards, or herbs that appear to be identical are actually different at the genetic level. An evolutionary interpretation of this data, then, demands that the morphologically identical organisms must have evolved independently of one another in a “repeatable” fashion.
Stephen J. Gould, Wonderful Life: The Burgess Shale and the Nature of History (1989), 51.
“…No finale can be specified at the start, none would ever occur a second time in the same way, because any pathway proceeds through thousands of improbable stages. Alter any early event, ever so slightly, and without apparent importance at the time, and evolution cascades into a radically different channel.1
Gould’s metaphor of “replaying life’s tape” asserts that if one were to push the rewind button, erase life’s history, and let the tape run again, the results would be completely different.2 The very essence of the evolutionary process renders evolutionary outcomes as nonreproducible (or nonrepeatable). Therefore, “repeatable” evolution is inconsistent with the mechanism available to bring about biological change.
Paleontologist J. William Schopf, one of the world’s leading authorities on early life on Earth, has made this very point in the book Life’s Origin.
Because biochemical systems comprise many intricately interlinked pieces, any particular full-blown system can only arise once…Since any complete biochemical system is far too elaborate to have evolved more than once in the history of life, it is safe to assume that microbes of the primal LCA cell line had the same traits that characterize all its present-day descendants.
"The nuclear pore in eukaryotes is a gigantic, complex structure with very distinctive ways of keeping most proteins out but specifically importing others. What we're probably looking at with the jumbo phage is a dramatically simpler method of solving the same problem," explains biochemist Kevin Corbett. "It's an amazingly creative solution – similar but simpler – to protecting its genome from the outside world by building a wall to separate it from bacterial defenses."
Incredibly, this shield can also grow as the phage's genome replicates. The researchers aren't quite sure how phages manage this yet but suspect the compartment is probably cracking open to allow more chimallin units to join it, which are produced in abundance on infection.
Meiying Gao (2017): Only 93 phages with genomes larger than 200 kbp have been isolated during the past 100 years since the discovery of phages (up to 30 June 2016). More than 80% of these were isolated during the past 3 years, which might be because of the revitalization of phage research and the progress in next-generation genome sequence technology in recent years. Tailed phages with genomes larger than 200 kbp are classified as “jumbo phages,” and phages of this kind usually harbor large virions. The jumbo phages are evolutionarily distant from the small genome phages. Despite the common features that differentiate them from smaller-genome phages, jumbo phages show more divergent characteristics among each other, such as low genome similarity, individual virion substructure, and different propagation mechanisms. 3
Thomas G. Laughli et.al.,(2022): Bacteria encode myriad defences that target the genomes of infecting bacteriophage, including restriction–modification and CRISPR–Cas systems. In response, one family of large bacteriophages uses a nucleus-like compartment to protect its replicating genomes by excluding host defence factors. The bacteriophage nuclear shell assembles primarily from one protein, which we name chimallin (ChmA). Chimallin self-assembles as a flexible sheet into closed micrometre-scale compartments. The architecture and assembly dynamics of the chimallin shell suggest mechanisms for its nucleation and growth, and its role as a scaffold for phage-encoded factors mediating macromolecular transport, cytoskeletal interactions, and viral maturation.
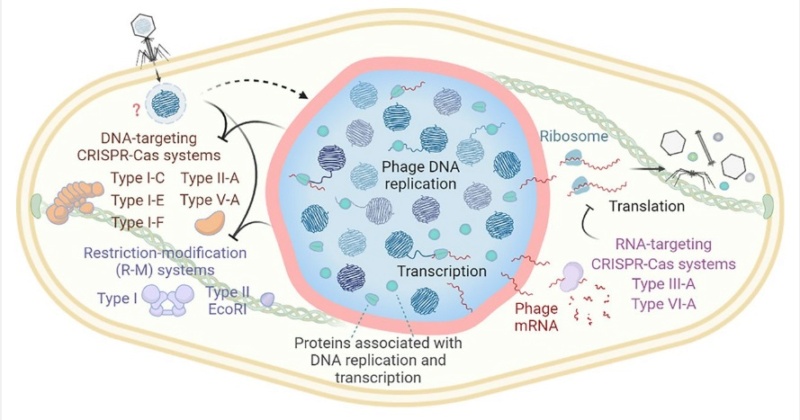
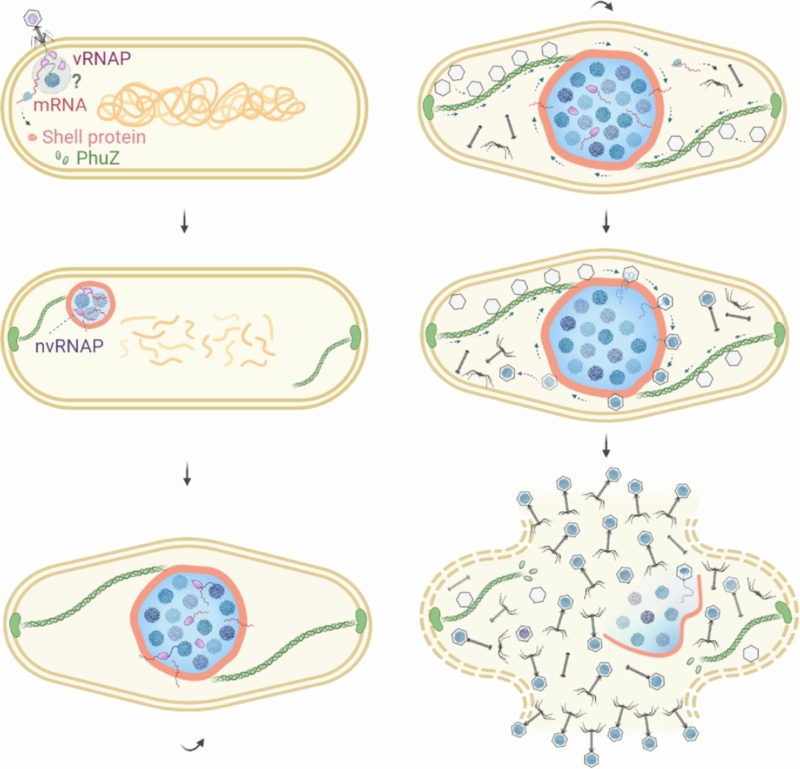
Bacteria have an array of defensive systems to target and destroy foreign nucleic acids. In turn, phages have mechanisms, including anti-restriction and anti-CRISPR proteins, that counter specific bacterial defence systems. A family of ‘jumbo phages’—named for their large genomes (typically over 200 kb) and large virion size assemble a selectively permeable, protein-based shell that encloses the replicating viral genome and is associated with a unique phage life cycle (Fig. 1a).
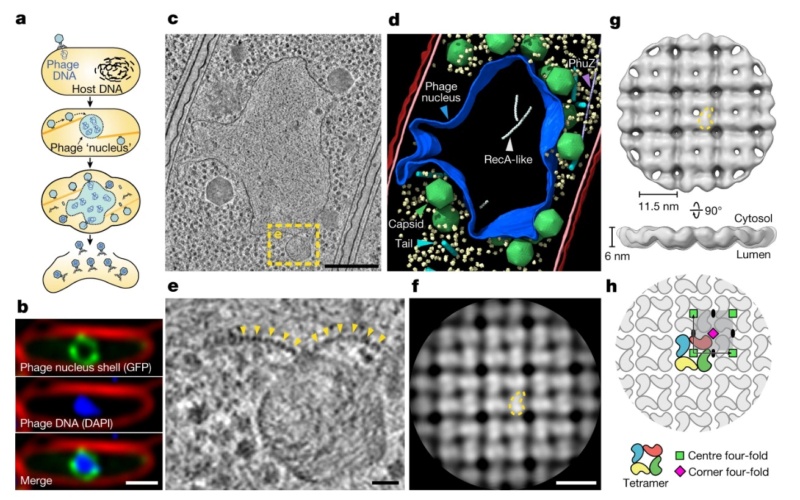
a, Schematic of the jumbo phage infection cycle.
b, Fluorescence microscopy of a 201phi2-1-infected P. chlororaphis cell at 45 mpi (n = 5 independent experiments). Phage nucleus shell component gp105 (green) is tagged with GFP, phage DNA (blue) is stained with DAPI and the outer cell membrane (red) is stained with FM4-64.
c, Tomographic slice of a phage nucleus in a 201phi2-1-infected P. chlororaphis cell at 50–60 mpi.
d, Segmentation of the tomogram in c. Outer and inner bacterial membranes are shown in burgundy and pink, respectively. The phage nucleus is coloured blue. Phage capsids and tails are green and cyan, respectively. PhuZ and RecA-like protein filaments are light purple and white, respectively. A subset of 500 host ribosomes is shown in pale yellow.
e, Enlarged view of the boxed region in c. Yellow arrows point to the repetitive feature of the phage nucleus perimeter.
f, Slice of the cytosolic face of the subtomogram average of the repetitive feature in the phage nucleus perimeter with a comma-shaped subunit outlined in yellow.
g, Cytosolic and side views of the shell subtomogram average isosurface with a single subunit outlined in yellow. h, Schematic representation of the p442-like arrangement of chimallin protomers. A ‘centre’ four-fold symmetry is indicated by a green square and a ‘corner’ four-fold symmetry is indicated by a magenta square. Scale bars: 1 μm (b), 250 nm (c), 25 nm (e) and 10 nm (f).
This micrometre-scale nucleus-like compartment, termed the ‘phage nucleus’, forms de novo upon infection and grows with the replicating viral DNA. Meanwhile, phage proteins are synthesized in the host cell cytoplasm. These include PhuZ, a phage-encoded tubulin homologue that assembles into filaments that treadmill to transport empty capsids from their assembly sites at the host cell membrane to the surface of the phage nucleus. Capsids dock to the phage nuclear shell and are filled with viral DNA before detaching and completing assembly with phage tails. Mature particles are released by host cell lysis. In contrast to other characterized anti-restriction systems, the phage nuclear shell renders jumbo phages broadly immune to DNA-targeting host restriction systems, including CRISPR–Cas, throughout infection by serving as a physical barrier between the viral DNA and host nucleases.
Chimallin is the principal shell protein
We previously showed that the abundant and early-expressed 201phi2-1 protein gp105 becomes integrated into the nuclear shell2. Furthermore, 201phi2-1 gp105 has a molecular weight of 69.5 kDa (631 amino acids). Along with the high apparent compositional homogeneity of the shell, these data support 201phi2-1 gp105 as the principal component of the phage nuclear shell. Homologues of 201phi2-1 gp105 are encoded by a large set of jumbo phage that infect diverse bacteria including Pseudomonas, Vibrio, Salmonella, and Escherichia coli, but these proteins bear no detectable sequence homology to any other proteins.
Comment: This is one of the certainly many examples showing the lack of sequence homology, and refutes Hong Zhou's (2015) claim that: Viruses likely originated from ancient cells through encapsulating cellular plasmids or genome fragments by cellular protein 1 Consider as well its size, 631 amino acids are composing a protein of considerable length and enormous sequence space.
Because of its role in protecting the phage genome against host defences, we named this protein chimallin (ChmA) after the chimalli, a shield carried by ancient Aztec warriors10. To understand the structure and assembly mechanisms of chimallin, we expressed and purified 201phi2-1 chimallin from E. coli. Size-exclusion chromatography coupled to multi-angle light scattering (SEC–MALS) of purified chimallin indicated a mixture of oligomeric states including monomers, well-defined assemblies of approximately 1.2 MDa, and larger heterogeneous species ranging from 4 to 13 MDa (mean of 6.9 MDa; Fig. 2a).
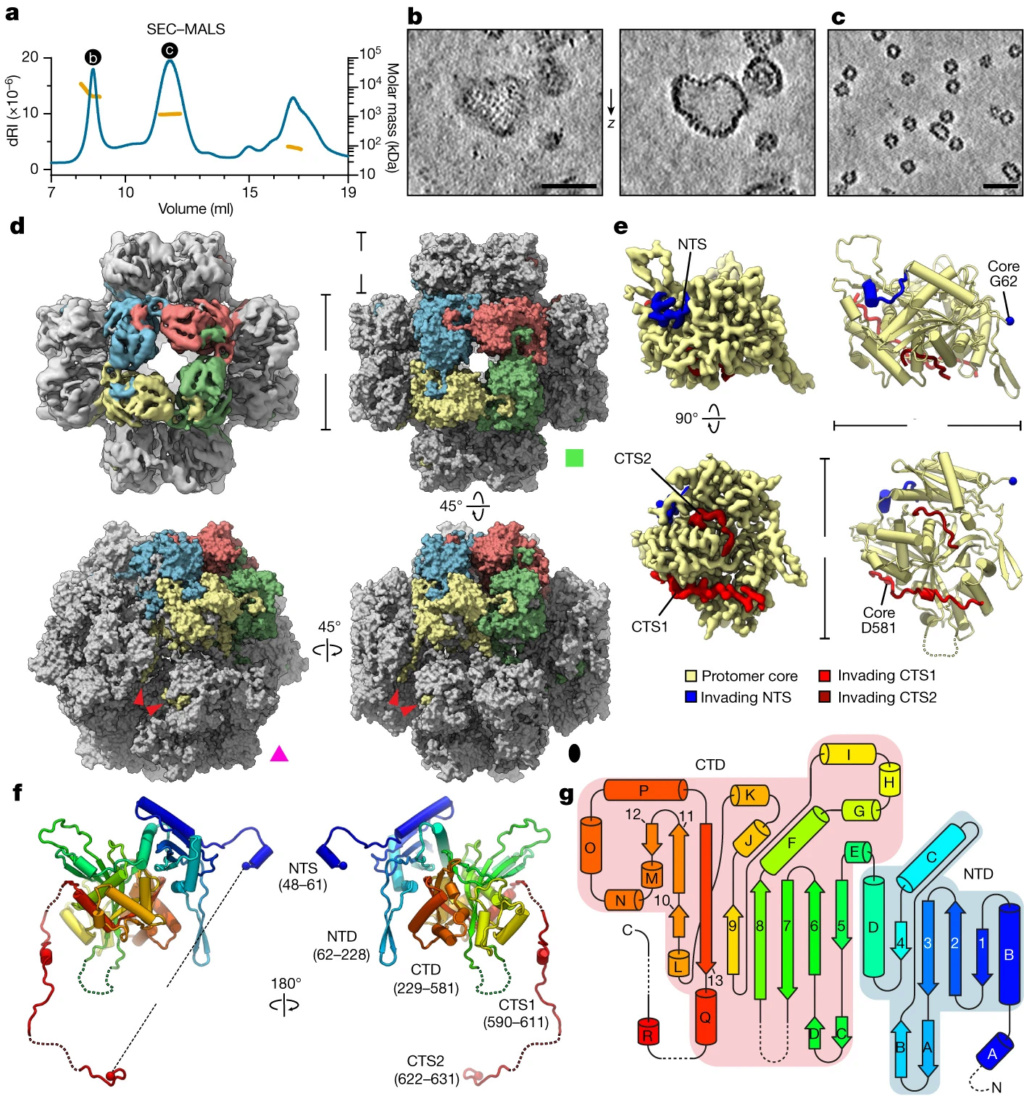
a, SEC–MALS analysis of purified 201phi2-1 chimallin. The measured molar masses of the three peaks are 6.9 MDa (range 4–13 MDa), 1.2 MDa and 87 kDa (left to right). dRI, differential refractive index.
b,c, Z-slices from tomograms of samples from the correspondingly labelled SEC–MALS peaks in a.
d, Top left, O-symmetrized reconstruction of the chimallin cubic assembly viewed along the four-fold axis. The protomers of one four-fold face are coloured. Top right, surface representation of the chimallin cubic assembly model viewed along the four-fold axis. Bottom right and bottom left, views of the model along the two-fold and three-fold axes, respectively. Red arrowheads point to the C-terminal segments of the yellow protomer. The green square, pink triangle and black oval indicate that the corresponding panels are viewed down the particle’s four-fold, three-fold and two-fold rotational symmetry axes, respectively.
e, Localized asymmetric reconstruction of the chimallin protomer (left) and cartoon model (right). Invading N- and C-terminal segments from neighbouring protomers are coloured blue (NTS), red (CTS1) and burgundy (CTS2). Resolved core protomer termini are shown as spheres.
f, Rainbow-coloured cartoon model of the chimallin protomer conformation in the cubic assembly. Resolved N and C termini are shown as spheres. Domains and segments are labelled. Unresolved linkers are shown as dashed lines.
g, A rainbow-coloured fold diagram of chimallin (blue at N terminus, red at C terminus) with α-helices labelled alphabetically and β-strands labelled numerically. The N- and C-terminal domains are highlighted in blue and red, respectively. Dashed lines indicate unresolved loops. Scale bars, 50 nm.
Cryo-ET of the largest species revealed pleomorphic, closed compartments with near-identical morphology to the phage nuclear shell that we observe in situ (Fig. 2b). Analysis of the smaller, more defined assemblies by cryo-ET revealed a near-homogeneous population of cubic assemblies with a diameter of around 22 nm, and a minor population of rectangular assemblies with dimensions of approximately 22 × 33.5 nm (Fig. 2c). 2
Giant Viruses Called 'Jumbo Phages' and chimallin, its principal shell protein
https://www.youtube.com/watch?v=zCs-sgcHWVc
Emerging Aspects of Jumbo Bacteriophages
1. TESSA KOUMOUNDOUROS Giant Viruses Called 'Jumbo Phages' Could Help Us Fight Antibiotic Resistance 05 August 2022
2. Thomas G. Laughlin: Architecture and self-assembly of the jumbo bacteriophage nuclear shell 03 August 2022
3. Meiying Gao Jumbo Bacteriophages: An Overview 14 March 2017
https://reasonandscience.catsboard.com/t3256-giant-viruses-called-jumbo-phages
TESSA KOUMOUNDOUROS (2022): As antibiotic-resistant bacteria become increasingly challenging to defeat, consulting those who've been in battle with our ancient foe long before us just makes a whole lot of sense. Viruses called bacteriophages have been clashing with bacteria since long before we even existed. So researchers from the University of California, San Diego, took a closer look at some of the lesser studied jumbo phages, specifically 201phi2-1, which infects Pseudomonas chlororaphis bacteria.
Named for their large genomes of over 200,000 base pairs long (most phages that infect Pseudomonas have genomes less than 100,000 base pairs), jumbo phages have an arsenal of techniques to counter bacterial defense mechanisms. Previously research found growing a shield around its genetic material was one of these tactics, making these viruses' life cycle unique.

The jumbo phage compared to T4 phage virus.
Separation of genomic material from the rest of the cell contents by encasing it "was previously thought to have evolved only once in the history of life", molecular biologist Thomas Laughlin and colleagues write in their new paper. But it turns out these tiny giants beat us to it.
"It's a different kind of compartment – unlike anything we have ever seen in nature," says biophysicist Elizabeth Villa.
Usually, phages inject their genetic material into the bacteria where it floats freely in the cell's inner goo (cytoplasm) as the virus hijacks the bacteria's equipment to replicate itself. But these jumbo phages construct a separating compartment around their DNA soon after it enters its host, a bit like how our cells have a nucleus to protect our DNA.
This physically prevents the bacteria's CRISPR 'immune system' and other defensive enzymes from messing with the viral DNA. Laughlin and their team used cryo-electron microscopy and tomography to examine this compartment down to the atomic scale. The protective casing was constructed from just a single type of protein, which the researchers called chimallin, after an ancient Aztec shield.

Micrograph of jumbo phage-infected bacterial cell (left) and nucleus-like compartment outlined in blue (right).
With the help of computer modeling, the researchers found the phage nucleus selectively allows molecules to pass through tiny pores – again similar to how cells control the environment around our genetic material, making this a remarkable example of convergent evolution – when completely unrelated organisms end up with a similar solution to the same biological problem.
Comment:
Convergence, another problem for evolution
https://reasonandscience.catsboard.com/t2014-convergence-another-problem-for-evolution
Biologists are uncovering numerous examples of organisms that cluster together morphologically (structurally), and yet are genetically distinct. Frogs, lizards, or herbs that appear to be identical are actually different at the genetic level. An evolutionary interpretation of this data, then, demands that the morphologically identical organisms must have evolved independently of one another in a “repeatable” fashion.
Stephen J. Gould, Wonderful Life: The Burgess Shale and the Nature of History (1989), 51.
“…No finale can be specified at the start, none would ever occur a second time in the same way, because any pathway proceeds through thousands of improbable stages. Alter any early event, ever so slightly, and without apparent importance at the time, and evolution cascades into a radically different channel.1
Gould’s metaphor of “replaying life’s tape” asserts that if one were to push the rewind button, erase life’s history, and let the tape run again, the results would be completely different.2 The very essence of the evolutionary process renders evolutionary outcomes as nonreproducible (or nonrepeatable). Therefore, “repeatable” evolution is inconsistent with the mechanism available to bring about biological change.
Paleontologist J. William Schopf, one of the world’s leading authorities on early life on Earth, has made this very point in the book Life’s Origin.
Because biochemical systems comprise many intricately interlinked pieces, any particular full-blown system can only arise once…Since any complete biochemical system is far too elaborate to have evolved more than once in the history of life, it is safe to assume that microbes of the primal LCA cell line had the same traits that characterize all its present-day descendants.
"The nuclear pore in eukaryotes is a gigantic, complex structure with very distinctive ways of keeping most proteins out but specifically importing others. What we're probably looking at with the jumbo phage is a dramatically simpler method of solving the same problem," explains biochemist Kevin Corbett. "It's an amazingly creative solution – similar but simpler – to protecting its genome from the outside world by building a wall to separate it from bacterial defenses."
Incredibly, this shield can also grow as the phage's genome replicates. The researchers aren't quite sure how phages manage this yet but suspect the compartment is probably cracking open to allow more chimallin units to join it, which are produced in abundance on infection.
Meiying Gao (2017): Only 93 phages with genomes larger than 200 kbp have been isolated during the past 100 years since the discovery of phages (up to 30 June 2016). More than 80% of these were isolated during the past 3 years, which might be because of the revitalization of phage research and the progress in next-generation genome sequence technology in recent years. Tailed phages with genomes larger than 200 kbp are classified as “jumbo phages,” and phages of this kind usually harbor large virions. The jumbo phages are evolutionarily distant from the small genome phages. Despite the common features that differentiate them from smaller-genome phages, jumbo phages show more divergent characteristics among each other, such as low genome similarity, individual virion substructure, and different propagation mechanisms. 3
Thomas G. Laughli et.al.,(2022): Bacteria encode myriad defences that target the genomes of infecting bacteriophage, including restriction–modification and CRISPR–Cas systems. In response, one family of large bacteriophages uses a nucleus-like compartment to protect its replicating genomes by excluding host defence factors. The bacteriophage nuclear shell assembles primarily from one protein, which we name chimallin (ChmA). Chimallin self-assembles as a flexible sheet into closed micrometre-scale compartments. The architecture and assembly dynamics of the chimallin shell suggest mechanisms for its nucleation and growth, and its role as a scaffold for phage-encoded factors mediating macromolecular transport, cytoskeletal interactions, and viral maturation.
Bacteria have an array of defensive systems to target and destroy foreign nucleic acids. In turn, phages have mechanisms, including anti-restriction and anti-CRISPR proteins, that counter specific bacterial defence systems. A family of ‘jumbo phages’—named for their large genomes (typically over 200 kb) and large virion size assemble a selectively permeable, protein-based shell that encloses the replicating viral genome and is associated with a unique phage life cycle (Fig. 1a).

a, Schematic of the jumbo phage infection cycle.
b, Fluorescence microscopy of a 201phi2-1-infected P. chlororaphis cell at 45 mpi (n = 5 independent experiments). Phage nucleus shell component gp105 (green) is tagged with GFP, phage DNA (blue) is stained with DAPI and the outer cell membrane (red) is stained with FM4-64.
c, Tomographic slice of a phage nucleus in a 201phi2-1-infected P. chlororaphis cell at 50–60 mpi.
d, Segmentation of the tomogram in c. Outer and inner bacterial membranes are shown in burgundy and pink, respectively. The phage nucleus is coloured blue. Phage capsids and tails are green and cyan, respectively. PhuZ and RecA-like protein filaments are light purple and white, respectively. A subset of 500 host ribosomes is shown in pale yellow.
e, Enlarged view of the boxed region in c. Yellow arrows point to the repetitive feature of the phage nucleus perimeter.
f, Slice of the cytosolic face of the subtomogram average of the repetitive feature in the phage nucleus perimeter with a comma-shaped subunit outlined in yellow.
g, Cytosolic and side views of the shell subtomogram average isosurface with a single subunit outlined in yellow. h, Schematic representation of the p442-like arrangement of chimallin protomers. A ‘centre’ four-fold symmetry is indicated by a green square and a ‘corner’ four-fold symmetry is indicated by a magenta square. Scale bars: 1 μm (b), 250 nm (c), 25 nm (e) and 10 nm (f).
This micrometre-scale nucleus-like compartment, termed the ‘phage nucleus’, forms de novo upon infection and grows with the replicating viral DNA. Meanwhile, phage proteins are synthesized in the host cell cytoplasm. These include PhuZ, a phage-encoded tubulin homologue that assembles into filaments that treadmill to transport empty capsids from their assembly sites at the host cell membrane to the surface of the phage nucleus. Capsids dock to the phage nuclear shell and are filled with viral DNA before detaching and completing assembly with phage tails. Mature particles are released by host cell lysis. In contrast to other characterized anti-restriction systems, the phage nuclear shell renders jumbo phages broadly immune to DNA-targeting host restriction systems, including CRISPR–Cas, throughout infection by serving as a physical barrier between the viral DNA and host nucleases.
Chimallin is the principal shell protein
We previously showed that the abundant and early-expressed 201phi2-1 protein gp105 becomes integrated into the nuclear shell2. Furthermore, 201phi2-1 gp105 has a molecular weight of 69.5 kDa (631 amino acids). Along with the high apparent compositional homogeneity of the shell, these data support 201phi2-1 gp105 as the principal component of the phage nuclear shell. Homologues of 201phi2-1 gp105 are encoded by a large set of jumbo phage that infect diverse bacteria including Pseudomonas, Vibrio, Salmonella, and Escherichia coli, but these proteins bear no detectable sequence homology to any other proteins.
Comment: This is one of the certainly many examples showing the lack of sequence homology, and refutes Hong Zhou's (2015) claim that: Viruses likely originated from ancient cells through encapsulating cellular plasmids or genome fragments by cellular protein 1 Consider as well its size, 631 amino acids are composing a protein of considerable length and enormous sequence space.
Because of its role in protecting the phage genome against host defences, we named this protein chimallin (ChmA) after the chimalli, a shield carried by ancient Aztec warriors10. To understand the structure and assembly mechanisms of chimallin, we expressed and purified 201phi2-1 chimallin from E. coli. Size-exclusion chromatography coupled to multi-angle light scattering (SEC–MALS) of purified chimallin indicated a mixture of oligomeric states including monomers, well-defined assemblies of approximately 1.2 MDa, and larger heterogeneous species ranging from 4 to 13 MDa (mean of 6.9 MDa; Fig. 2a).

a, SEC–MALS analysis of purified 201phi2-1 chimallin. The measured molar masses of the three peaks are 6.9 MDa (range 4–13 MDa), 1.2 MDa and 87 kDa (left to right). dRI, differential refractive index.
b,c, Z-slices from tomograms of samples from the correspondingly labelled SEC–MALS peaks in a.
d, Top left, O-symmetrized reconstruction of the chimallin cubic assembly viewed along the four-fold axis. The protomers of one four-fold face are coloured. Top right, surface representation of the chimallin cubic assembly model viewed along the four-fold axis. Bottom right and bottom left, views of the model along the two-fold and three-fold axes, respectively. Red arrowheads point to the C-terminal segments of the yellow protomer. The green square, pink triangle and black oval indicate that the corresponding panels are viewed down the particle’s four-fold, three-fold and two-fold rotational symmetry axes, respectively.
e, Localized asymmetric reconstruction of the chimallin protomer (left) and cartoon model (right). Invading N- and C-terminal segments from neighbouring protomers are coloured blue (NTS), red (CTS1) and burgundy (CTS2). Resolved core protomer termini are shown as spheres.
f, Rainbow-coloured cartoon model of the chimallin protomer conformation in the cubic assembly. Resolved N and C termini are shown as spheres. Domains and segments are labelled. Unresolved linkers are shown as dashed lines.
g, A rainbow-coloured fold diagram of chimallin (blue at N terminus, red at C terminus) with α-helices labelled alphabetically and β-strands labelled numerically. The N- and C-terminal domains are highlighted in blue and red, respectively. Dashed lines indicate unresolved loops. Scale bars, 50 nm.
Cryo-ET of the largest species revealed pleomorphic, closed compartments with near-identical morphology to the phage nuclear shell that we observe in situ (Fig. 2b). Analysis of the smaller, more defined assemblies by cryo-ET revealed a near-homogeneous population of cubic assemblies with a diameter of around 22 nm, and a minor population of rectangular assemblies with dimensions of approximately 22 × 33.5 nm (Fig. 2c). 2
Giant Viruses Called 'Jumbo Phages' and chimallin, its principal shell protein
https://www.youtube.com/watch?v=zCs-sgcHWVc
Emerging Aspects of Jumbo Bacteriophages
1. TESSA KOUMOUNDOUROS Giant Viruses Called 'Jumbo Phages' Could Help Us Fight Antibiotic Resistance 05 August 2022
2. Thomas G. Laughlin: Architecture and self-assembly of the jumbo bacteriophage nuclear shell 03 August 2022
3. Meiying Gao Jumbo Bacteriophages: An Overview 14 March 2017
Last edited by Otangelo on Thu Sep 22, 2022 10:20 am; edited 1 time in total