The pan-immune system of bacteria: antiviral defense as a community resource
Aude Bernheim (2019): Bacteriophages (phages) are the most abundant viruses on the planet. The majority of free-living bacterial species are thought to be infected by phages, as evidenced by the widespread presence of prophages (dormant phages) in bacterial genomes. The arms race between bacteria and phages are considered almost as old as bacteria themselves. Facing the abundance and diversity of phages, bacteria have developed multiple lines of defense that can collectively be referred to as the ‘prokaryotic immune system’. In recent years, it has been recognized that prokaryotic immunity is much more complex than previously perceived, with evidence for chemical defense and intracellular signalling regulating defence that are relevant also for archaeal defense systems that protect from archaeal viruses. Diversity of defense systems Anti-phage defense systems can roughly be divided into those that target viral nucleic acids (for example, R-M and CRISPR–Cas), Abi systems that lead the host to commit suicide once infected, and other types of systems (Fig. 1).
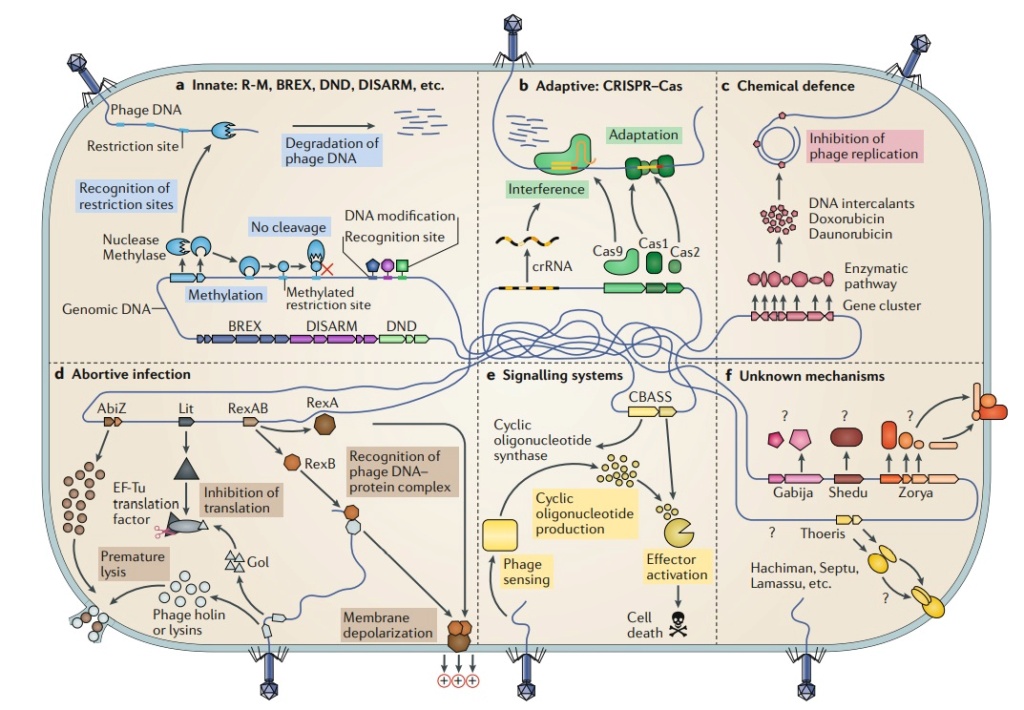
Figure 1. Antiviral defense systems in bacteria.
Defence systems that target nucleic acids encompass both innate and adaptive immunity.
a | Restriction-modification (R-M) and other related systems modify specific sequence motifs in the host genome and cleave or degrade unmodified foreign DNA.
b | CRISPR–Cas systems work in two main phases: adaptation, where a complex of Cas proteins guides the acquisition of new bacteriophage (phage)-derived spacers; and interference, where Cas proteins in a complex with a spacer-derived CRISPR RNA (crRNA) target and degrade phage nucleic acids.
c | Chemical defense has been described in Streptomyces spp., in which bacteria produce a small anti-phage molecule that intercalates into phage DNA and inhibits its replication.
d | Abortive infection mechanisms are diverse. In concert with phage-encoded holins and lysins of phage Phi31, AbiZ from Lactococcus lactis accelerates lysis before phage assembly is completed. Upon expression of the T4 phage protein Gol, the Escherichia coli Lit protein inhibits translation through cleavage of the EF-Tu elongation factor. The E. coli protein RexA recognizes a specific DNA–protein complex formed by the λ phage, and activates RexB, an ion channel that depolarizes the membrane, leading to cell death.
e | CBASS (cyclic oligonucleotide-based anti-phage signalling system) senses the presence of phage and generates a cyclic oligonucleotide small-molecule signal that activates an effector leading to cell death.
f | Multiple systems have recently been demonstrated to have anti-phage roles, but their mechanisms remain unknown. Abi, abortive infection; BREX, bacteriophage exclusion; DISARM, defence islands system associated with R-M.
Of these, the most abundant and elaborate systems are those that target nucleic acids, presumably because nucleic acid is usually the first viral component to penetrate the cell upon infection (Fig. 1a,b). R-M collectively refers to systems that cleave or degrade DNA through the recognition of specific sequence motifs on the viral genome. These sequence motifs are modified in the host self-DNA, usually by methylation, to prevent the host genome from being targeted (with the exception of type IV R-M systems, which target modified phage DNA while the host genome remains unaltered). R-M systems are classified into four types and are present in more than 74% of prokaryotic genomes. On average, a bacterial genome encodes two R-M systems. DNA modification as a strategy to discriminate between self-DNA and non-self-DNA is not limited to methylation. For example, the dnd defense system modifies the host DNA backbone to include a sulfur group, and the dpd system utilizes a multi-enzyme pathway to modify guanine residues into 7-deazaguanine derivatives in the host DNA. The BREX (bacteriophage exclusion) system and DISARM (defense islands system associated with R-M) also function through methylation of host DNA, although the mechanisms of phage DNA targeting in these systems are still unknown. All of these defense systems constitute part of the innate immunity of bacteria. A large fraction of bacteria and archaea encode CRISPR–Cas1, a family of adaptive immune systems that also function through the recognition and degradation of viral nucleic acids. The CRISPR–Cas immune memory is formed through the acquisition of short viral-derived DNA sequences that are incorporated as CRISPR ‘spacers’ within the outcome, as well as the discovery of a large number of new defense systems whose mechanisms are still unknown. Individual bacterial species can encode multiple different defense systems, and it was shown that such systems can be horizontally acquired and lost on short evolutionary time scales.
Diversity of defense systems Anti-phage defense systems can roughly be divided into those that target viral nucleic acids (for example, R-M and CRISPR–Cas), Abi systems that lead the host to commit suicide once infected, and other types of systems (Fig. 1). Of these, the most abundant and elaborate systems are those that target nucleic acids, presumably because nucleic acid is usually the first viral component to penetrate the cell upon infection (Fig. 1a,b). R-M collectively refers to systems that cleave or degrade DNA through recognition of specific sequence motifs on the viral genome. These sequence motifs are modified in the host self-DNA, usually by methylation, to prevent the host genome from being targeted (with the exception of type IV R-M systems, which target modified phage DNA while the host genome remains unaltered). R-M systems are classified into four types and are present in more than 74% of prokaryotic genomes. On average, a bacterial genome encodes two R-M systems. DNA modification as a strategy to discriminate between self-DNA and non-self-DNA is not limited to methylation. For example, the dnd defense system modifies the host DNA backbone to include a sulfur group, and the dpd system utilizes a multi-enzyme pathway to modify guanine residues into 7-deazaguanine derivatives in the host DNA. The BREX (bacteriophage exclusion) system and DISARM (defence islands system associated with R-M) also function through methylation of host DNA, although the mechanisms of phage DNA targeting in these systems are still unknown. All of these defence systems constitute part of the innate immunity of bacteria. A large fraction of bacteria and archaea encode CRISPR–Cas, a family of adaptive immune systems that also function through the recognition and degradation of viral nucleic acids. The CRISPR–Cas immune memory is formed through acquisition of short viral-derived DNA sequences that are incorporated as CRISPR ‘spacers’ within the host genome. These sequences are then transcribed and processed into CRISPR RNAs that guide the CRISPR–Cas machinery, through sequence complementarity, to target the viral nucleic acids. CRISPR–Cas systems are diverse, comprising two classes, six types and more than 20 subtypes that differ in the composition of the interference machinery, their mechanisms of targeting and the nucleic acid targeted (that is, DNA or RNA). In most cases, both spacer acquisition and interference necessitate the occurrence of a short sequence motif named PAM (protospacer adjacent motif) next to the sequence matched by the spacer in the targeted molecule. Operons that include prokaryotic argonautes have also been hypothesized to provide defense. Present in 9% and 32% of bacterial and archaeal genomes, respectively, their frequent localization in defense islands (regions in microbial genomes in which defense systems are concentrated) as well as their protective activity against plasmids suggest that they are involved in antiviral defense. Another common strategy of defense against phages is Abi. Abi systems allow the bacterial cell, once infected, to kill itself or to arrest its metabolism before the phage reproductive cycle is completed, thus preventing the phage from spreading and killing the surrounding bacterial community. Abi systems have been detected in a wide variety of microorganisms but, given their high diversity, it is challenging to assess their abundance in nature. These systems are usually triggered by a specific component that could be a phage protein, a nucleic acid or a cellular state caused by phage infection. For example, the Escherichia coli Lit Abi is activated upon sensing a unique substrate formed by the Gol peptide of phage T4 when bound to the ribosomal elongation factor EF-Tu. Once active, the Lit protein cleaves EF-Tu, thus inhibiting translation and ultimately killing the cell.

Box 1: Antiviral defense systems tend to cluster on bacterial and archaeal chromosomes in regions denoted as defense islands. Defense islands typically comprise diverse defense systems. see the figure for examples of defense islands within selected bacterial and archaeal genomes (different colors represent different defense systems). Beige represents genes of non-defense functions or of unknown function. Defense systems are also enriched with genes typical of mobile genetic elements such as transposases, recombinases, and conjugation genes. Some defense islands were predicted to encode more than 100 defense genes. The origin and the mechanism of formation of defense islands are currently unknown but could reflect different effects. First, co-localization of defense genes with mobile genes could facilitate the horizontal transfer of multiple defense systems from one bacterium to another in a single transfer event. Alternatively, such islands can be hotspots for integration of horizontally acquired genes, with defense systems clustering in defense islands through the ‘garbage and pile effect’, in which high rates of acquisition and loss are not strongly deleterious. in addition, such co-localization of defense genes could reflect functional links between the defense systems, including possible co-regulation or positive epistasis. the phenomenon of defense islands in bacterial genomes allows the prediction of novel defense systems through a ‘guilt by association approach. in this approach, protein families with unknown functions that are enriched in defense islands can be predicted to constitute new defense systems. This methodology has led to the discovery of individual defense systems such as BreX (bacteriophage exclusion) or DisarM (defense islands system associated with r-M), and its application in a systematic manner recently revealed nine new antiviral systems that are widespread in bacteria and archaea. For r-M and CrisPr–Cas systems, the type is indicated in parentheses.
Another example is PrrC in E. coli, which cleaves bacterial tRNALys molecules when it senses that the phage has suppressed bacterial R-M systems. In Lactococcus spp., many Abi genes (around 20) have been described: for example, AbiZ accelerates lysis before phage assembly whereas AbiB leads to non-specific degradation of mRNAs. In Staphylococcus spp., the serine threonine kinase STK2 protein is activated when exposed to the phage protein PacK, leading to phosphorylation of proteins involved in multiple cellular pathways and eventual cell death. Toxin–antitoxin systems, representing a large family of two-gene modules each comprising a toxin and an immunity component, were also shown to execute Abi in some cases, although their general role in defense against phages is still disputed. A newly discovered system called CBASS (cyclic oligonucleotide-based anti-phage signalling system) employs a specific form of Abi34. This system, appearing in more than 10% of sequenced bacterial genomes, relies on cyclic oligonucleotide signalling to provide defense against phages. Sensing of phage infection leads to the production of a cyclic oligonucleotide, for example cyclic GMP–AMP, which activates a downstream effector that causes cell death. The CBASS system is considered the prokaryotic ancestor of the cGAS–STING antiviral pathway in animals, which similarly relies on cyclic GMP–AMP signalling. Recent studies have revealed the existence of many additional families of antiviral defense systems in bacteria and archaea. An effort to map microbial defense islands (Box 1) has resulted in the discovery of nine new defense systems that are widespread in bacterial and archaeal genomes. These systems were named after protective deities from world mythologies including Hachiman, Thoeris, Zorya, Gabija and Shedu, and their molecular mechanisms of action are yet to be deciphered. Finally, species of Streptomyces produce small molecules called doxorubicin and daunorubicin that act as DNA intercalants, and were recently shown to specifically block phage DNA replication but not the replication of bacterial DNA4 .
A need for multiple defences
Analysis of sequenced prokaryotic genomes demonstrates that they can concomitantly harbour multiple different defense systems. As shown in Fig. 2, a single strain can encode diverse defence strategies including Abi, R-M and CRISPR–Cas. Many bacteria and archaea encode multiple defence systems of the same kind: for example, Helicobacter pylori F30 encodes three type I R-M systems, 11 type II R-M systems, one type III R-M system and one type IV R-M system15. In total, it was estimated that up to 10% of some prokaryotic genomes is dedicated to defence systems8 . These observations raise a basic question — what is the benefit for a single microorganism to encode so many different lines of defence?
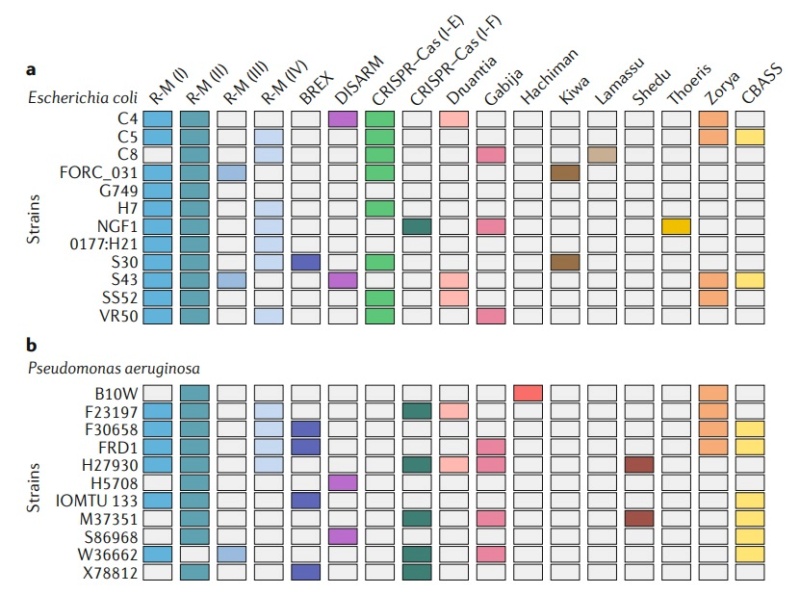
Fig. 2 | closely related bacterial strains encode diverse defense systems. Each line represents a different strain of either Escherichia coli (part a) or Pseudomonas aeruginosa (part b). Each column corresponds to a different defense system (color indicates the presence of a defense system). CRISPR–Cas systems were detected using CRISPRCasFinder, restriction-modification (R-M) systems using HHsearch with HMM profiles, BREX (bacteriophage exclusion) by the presence of the pglZ gene, DISARM (defense islands system associated with R-M) and CBASS (cyclic oligonucleotide-based anti-phage signalling system). For R-M and CRISPR–Cas systems, the type or subtype is indicated in parentheses. R-M, restriction-modification.
One obvious answer is that some defence systems can protect only from a specific type of virus. For example, the GmrSD type IV R-M system only targets phages such as T4, whose genomes are modified to include glucosylated hydroxymethylcytosine. Cas9, on the other hand, cannot cleave the DNA of phage T4 owing to its heavily modified cytosine residues. The Thoeris defence system seems to protect only against phages from the Myoviridae family . Therefore, for a microorganism to be protected against a wide variety of viruses, it should encode a broad defence arsenal that can overcome the multiple types of viruses that can infect it. There are benefits for a microorganism to encode multiple defense systems, even if these systems overlap in the range of viruses that they target. This is because phages can develop resistance to defense. First, phage genomes can evolve to eliminate specific sequences such as motifs targeted by restriction enzymes or PAM sequences that are essential for CRISPR–Cas defense. Second, phages often encode anti-defense proteins, including anti-CRISPR and anti-restriction proteins. These proteins are either injected into the cell together with the viral DNA or expressed early upon infection, and inhibit the defense systems. Anti-CRISPRs are typically short proteins that bind the CRISPR–Cas complex and prevent it from working properly. Recent discoveries report on anti-CRISPRs working as enzymes that can cleave the CRISPR RNA or add an acetyl group to a PAM-sensing residue in the Cas effector. Similarly, anti-restriction proteins inhibit restriction enzymes: for example, the T4 IPI (internal protein I) inhibits type IV R-M systems, whereas the DarA and DarB proteins of phage P1 bind the restriction sites on the phage genome and mask them from cleavage by the type I R-M system of E. coli. Faced with viruses that encode counter-defense mechanisms, bacteria and archaea cannot rely on a single defense system and thus need to present several lines of defense as a bet-hedging strategy of survival.
Gain and loss of defense systems
Owing to the selective advantage that defense systems provide, they are frequently gained by bacteria and archaea through horizontal gene transfer (HGT). Multiple studies based on phylogenetic analyses and comparative genomics have confirmed the high rate of transfer of defense systems. For example, only ∼4% of R-M systems are found in the core genomes of prokaryotic species, suggesting recent transfer events. In another example, an analysis of phylogenetic trees of Cas proteins and CRISPR repeats showed weak consistency with the species tree, demonstrating the dominance of horizontal transfer for the spread of CRISPR–Cas loci. Both CRISPR–Cas and R-M systems have been detected on mobile genetic elements, such as plasmids, transposons and phages, partially explaining their mode of HGT. In addition, genomic analyses have shown that defense systems tend to be concentrated in ‘defense islands’ — regions of the host chromosome that are also enriched with mobile elements presumably responsible for the genetic mobilization of the islands (Box 1). Given their selective advantage in the arms race against viruses, one might expect that defense systems, once acquired (either through direct evolution or via HGT), would accumulate in prokaryotic genomes and be selected for. Surprisingly, this is not the case as defense systems are known to be frequently lost from microbial genomes over short evolutionary time scales, suggesting that they can impose selective disadvantages in the absence of infection pressure. A major drawback of defense systems is autoimmunity: CRISPR–Cas, for example, can make mistakes in the process of spacer acquisition and acquire spacers from the chromosome instead of from the invading element. This directs the CRISPR–Cas interference machinery to attack the chromosome, resulting in cell death or in survival through pseudogenization and eventual deletion of the CRISPR–Cas locus. Similarly, R-M systems can also rarely target the chromosome, cleaving self-DNA at a low but measurable rate and inflicting a fitness cost. Unwanted activity of Abi systems can also lead to dormancy or cell death. In addition to autoimmunity, defense systems can impose an energy burden on the cell: some R-M systems require the hydrolysis of one ATP molecule per base pair for translocation of the restriction enzyme along the DNA. As a result of these fitness costs, there is a selective pressure for bacteria to get rid of defense systems under conditions when there is no selection pressure exerted by phages. Indeed, competition studies between strains encoding defense systems, such as CRISPR–Cas or Lit Abi, and cognate defense-lacking strains have demonstrated the existence of a fitness cost in the absence of phage infection. An experimental study in Staphylococcus epidermidis showed that the loss of CRISPR–Cas systems by large deletions have little or no fitness cost. Another study demonstrated that the inactivation of CRISPR–Cas systems in Streptococcus pneumoniae is even advantageous under specific conditions. The frequent gain and loss of defense systems over short time scales lead to a highly variable pattern of presence and absence of systems in microbial genomes. Even in closely related strains with otherwise similar genomes, the composition of defense systems can drastically vary, as demonstrated in Fig. 2. Defence systems appear to be in a state of constant genetic flux, constituting the second most dynamic class of genes after mobile genetic elements in terms of rates of gain and loss in microbial genomes.
Pan-immunity as a shared resource
Given the fitness costs inflicted by antiviral systems, it is probable that no single bacterial or archaeal strain can encode, in the long term, all possible defense systems without suffering serious competitive disadvantages. On the other hand, access to a diverse set of defense mechanisms is essential in order to combat the enormous genetic and functional diversity of viruses. We propose that these seemingly contradictory requirements can be reconciled when considering the available arsenal of immune systems as a resource shared by a population of bacteria or archaea rather than by individual cells. In the example shown in Fig. 2, none of the strains encode all defense systems. However, if these strains are mixed as part of a population, the pan-genome of this population would encode an ‘immune potential’ that encompasses all of the depicted systems. As these systems can be readily available by HGT, given the high rate of HGT in defense systems, the population in effect harbors an accessible reservoir of immune systems that can be acquired by population members. When the population is subjected to infection, this diversity ensures that at least some population members would encode the appropriate defense system, and these members would survive and form the basis for the perpetuation of the population (Fig. 3).
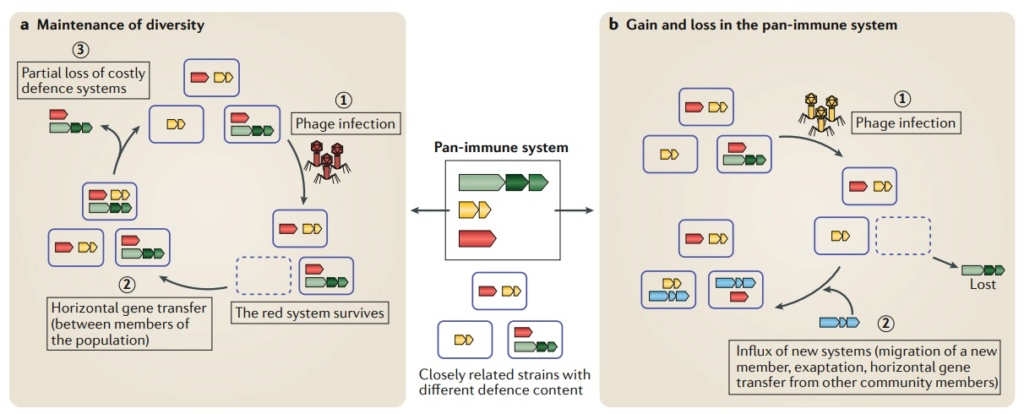
The pan-immune system model.
Closely related strains of microorganisms within a population encode a diverse set of antiviral systems constituting the pan-immune system defense.
a | Maintenance of diversity of the pan-immune system. Bacteriophage (phage) infection results in bacterial selection for those encoding a specific defense system (red) that can overcome that phage (part a, stage 1). In the absence of phage pressure over a period of time, members of the population can acquire a diverse set of defense systems, through horizontal gene transfer (HGT) (part a, stage 2), whereas some cells lose defense systems owing to their selective cost (part a, stage 3). The cycle continues, resulting in a population defense that together constitutes the immune potential of the population.
b | Dynamic changes to the pan-immune system composition. Phage infection results in selection for members encoding a specific defense system (yellow) that can overcome that phage (part b, stage 1). In some cases, this can result in the loss of immune system defense (green). Conversely, new systems can be introduced into the population through HGT from a more distantly related strain, migration of a new member in the population or emergence of a new system through mutation or exaptation (part b, stage 2). Red, green, yellow and blue genes represent different types of defense systems.
We thus hypothesize that some of the selection for defense systems occurs at the group level. In a sense, this pan-immune system model aligns well with previous observations and mathematical models of distributed immunity that specifically focused on CRISPR–Cas systems. Studies on CRISPR–Cas have shown that spacer diversity in the population is essential to overcoming phage infections. In co-evolution experiments between Pseudomonas aeruginosa and Streptococcus thermophilus and their respective phages, bacterial populations in which different strains encoded different sets of spacers overcame phage infection and resulted in phage extinction, whereas populations comprising homogeneous sets of spacers allowed phage propagation. Protection of spacer-diverse populations occurred because no single phage could accumulate enough mutations to overcome the diversity of spacers encoded by the population as a whole. In the context of CRISPR–Cas, mathematical models that explored the parameters leading to the emergence of a distributed immunity predicted two key parameters: the cost of generating a new allele (in this case, a new spacer) should be small; and the fitness constraints of evolving escape mutations for phages is enhanced by the fact that an escape mutant will be resistant only to one allele (one spacer in the case of the CRISPR model). Beyond the specific case of CRISPR–Cas, the same conditions also fit the broader context of the microbial pan-immune system model, which can be viewed as satisfying the two parameters mentioned above: given the high rate of HGT of defense systems (which can be considered the acquisition of alleles of defense), the cost of acquiring a new allele via HGT is expected to be relatively small; and due to the diversity of molecular mechanisms among different defense systems, the emergence of one phage mutation that allows escape from a specific defense system is not expected to abolish defense by other systems. As group selection occurs within closely related kin, we expect the pan-immune system model to be mainly relevant among populations of similar, related strains that differ in their defense content, thus allowing for selection at the group level.
Implications for counter-defence
It is well documented that individual phages have well-defined host ranges, such that they can infect some, but rarely all, strains of the same species. This is often attributed to the diversity of surface molecules among the infected microbial strains, as these are used by phages as specific receptors. However, given the diversity of defense systems observed in different strains of the same species, it is clear that the host range of any given phage would depend on its ability to overcome multiple defence systems. This predicts that phages need to encode many different counter-defense mechanisms in order to have a broad host range. This prediction may help reconcile the puzzle of dispensable genes in phage genomes. As phage genomes are under strong selection, one might expect that most of their genes are essential. However, serial mutational analyses showed that as much as 79% of genes in phage T4 and 63% of genes in phage T7 are not essential for successful infection of the E. coli laboratory strain. We predict that many of these genes will turn out to encode anti-defense proteins that target defense systems not present in the E. coli strain used in these studies. We would therefore expect that the set of anti-defense genes cumulatively encoded by strains of a phage species should mirror the set of defense systems encoded by its host pan-genome. 1
Evolution of Immune Systems From Viruses and Transposable Elements
Felix Broecker (2019): Cellular organisms have co-evolved with various mobile genetic elements (MGEs), including transposable elements (TEs), retroelements, and viruses, many of which can integrate into the host DNA. MGEs constitute ∼50% of mammalian genomes, >70% of some plant genomes, and up to 30% of bacterial genomes. 2
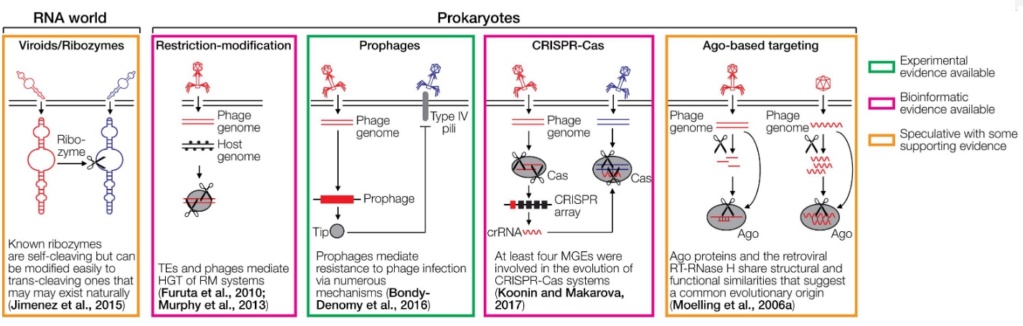
FIGURE 1. Cartoons depicting various defense systems.
The systems are color-coded based on the level of support in green (experimental evidence available), magenta (bioinformatic evidence available), and orange (speculative with some supporting evidence). DNA/RNA cleaving is indicated with scissors. RNA is depicted as wavy lines or with secondary hairpin-loop structures. A ribozyme cleaving another ribozyme is a hypothetical early immune system that does not require DNA or proteins. The ribozyme may be part of a viroid-like selfish RNA. Restriction-modification systems distinguish between foreign and self DNA by methylating target sequences of restriction endonucleases. Prophages can mediate superinfection exclusion, exemplified by the expression of the Tip protein that reduces bacterial surface expression of type IV pili required for infection by various phages. CRISPR-Cas acts by incorporating small genomic fragments from phages into CRISPR arrays in the prokaryotic genome. The transcribed spacers are then used by another Cas member to cleave sequence-homologous phage genomes. PIWI-associated RNAs (piRNAs) are small RNAs complementary to transposable elements (TEs) that are encoded in piRNA clusters. piRNAs associate with a PIWI nuclease to cleave complementary TE transcripts. RNA interference (RNAi) is initiated by dsRNA which is fragmented by Dicer to siRNAs. These siRNAs are loaded into the RNA-induced silencing complex to cleave complementary RNAs using Ago nucleases. A variation of RNAi is the endo-siRNA pathway, in which dsRNA is generated from TEs that are transcribed in both orientations, for instance, if the TE is located in an intron in opposite orientation to the encompassing gene. Endogenous retroviruses (ERVs) can mediate the restriction of ERVs and exogenous retroviruses through various mechanisms, including receptor blockade by captured Env proteins, Gag-mediated restriction, and antisense RNA mechanisms. The interferon system recognizes dsRNA or other pathogen-associated molecular patterns, which leads to the upregulation of antiviral interferon-stimulated genes (ISGs). The antibody system involves diversification through light and heavy chain recombination, which is mediated by the Rag recombinases. This enables the detection of diverse pathogens. Note that the references provided in the figure are not comprehensive. Please refer to the text for more details and additional references.
1. Aude Bernheim: The pan-immune system of bacteria: antiviral defence as a community resource 06 November 2019
2. Felix Broecker: Evolution of Immune Systems From Viruses and Transposable Elements 29 January 2019
Aude Bernheim (2019): Bacteriophages (phages) are the most abundant viruses on the planet. The majority of free-living bacterial species are thought to be infected by phages, as evidenced by the widespread presence of prophages (dormant phages) in bacterial genomes. The arms race between bacteria and phages are considered almost as old as bacteria themselves. Facing the abundance and diversity of phages, bacteria have developed multiple lines of defense that can collectively be referred to as the ‘prokaryotic immune system’. In recent years, it has been recognized that prokaryotic immunity is much more complex than previously perceived, with evidence for chemical defense and intracellular signalling regulating defence that are relevant also for archaeal defense systems that protect from archaeal viruses. Diversity of defense systems Anti-phage defense systems can roughly be divided into those that target viral nucleic acids (for example, R-M and CRISPR–Cas), Abi systems that lead the host to commit suicide once infected, and other types of systems (Fig. 1).

Figure 1. Antiviral defense systems in bacteria.
Defence systems that target nucleic acids encompass both innate and adaptive immunity.
a | Restriction-modification (R-M) and other related systems modify specific sequence motifs in the host genome and cleave or degrade unmodified foreign DNA.
b | CRISPR–Cas systems work in two main phases: adaptation, where a complex of Cas proteins guides the acquisition of new bacteriophage (phage)-derived spacers; and interference, where Cas proteins in a complex with a spacer-derived CRISPR RNA (crRNA) target and degrade phage nucleic acids.
c | Chemical defense has been described in Streptomyces spp., in which bacteria produce a small anti-phage molecule that intercalates into phage DNA and inhibits its replication.
d | Abortive infection mechanisms are diverse. In concert with phage-encoded holins and lysins of phage Phi31, AbiZ from Lactococcus lactis accelerates lysis before phage assembly is completed. Upon expression of the T4 phage protein Gol, the Escherichia coli Lit protein inhibits translation through cleavage of the EF-Tu elongation factor. The E. coli protein RexA recognizes a specific DNA–protein complex formed by the λ phage, and activates RexB, an ion channel that depolarizes the membrane, leading to cell death.
e | CBASS (cyclic oligonucleotide-based anti-phage signalling system) senses the presence of phage and generates a cyclic oligonucleotide small-molecule signal that activates an effector leading to cell death.
f | Multiple systems have recently been demonstrated to have anti-phage roles, but their mechanisms remain unknown. Abi, abortive infection; BREX, bacteriophage exclusion; DISARM, defence islands system associated with R-M.
Of these, the most abundant and elaborate systems are those that target nucleic acids, presumably because nucleic acid is usually the first viral component to penetrate the cell upon infection (Fig. 1a,b). R-M collectively refers to systems that cleave or degrade DNA through the recognition of specific sequence motifs on the viral genome. These sequence motifs are modified in the host self-DNA, usually by methylation, to prevent the host genome from being targeted (with the exception of type IV R-M systems, which target modified phage DNA while the host genome remains unaltered). R-M systems are classified into four types and are present in more than 74% of prokaryotic genomes. On average, a bacterial genome encodes two R-M systems. DNA modification as a strategy to discriminate between self-DNA and non-self-DNA is not limited to methylation. For example, the dnd defense system modifies the host DNA backbone to include a sulfur group, and the dpd system utilizes a multi-enzyme pathway to modify guanine residues into 7-deazaguanine derivatives in the host DNA. The BREX (bacteriophage exclusion) system and DISARM (defense islands system associated with R-M) also function through methylation of host DNA, although the mechanisms of phage DNA targeting in these systems are still unknown. All of these defense systems constitute part of the innate immunity of bacteria. A large fraction of bacteria and archaea encode CRISPR–Cas1, a family of adaptive immune systems that also function through the recognition and degradation of viral nucleic acids. The CRISPR–Cas immune memory is formed through the acquisition of short viral-derived DNA sequences that are incorporated as CRISPR ‘spacers’ within the outcome, as well as the discovery of a large number of new defense systems whose mechanisms are still unknown. Individual bacterial species can encode multiple different defense systems, and it was shown that such systems can be horizontally acquired and lost on short evolutionary time scales.
Diversity of defense systems Anti-phage defense systems can roughly be divided into those that target viral nucleic acids (for example, R-M and CRISPR–Cas), Abi systems that lead the host to commit suicide once infected, and other types of systems (Fig. 1). Of these, the most abundant and elaborate systems are those that target nucleic acids, presumably because nucleic acid is usually the first viral component to penetrate the cell upon infection (Fig. 1a,b). R-M collectively refers to systems that cleave or degrade DNA through recognition of specific sequence motifs on the viral genome. These sequence motifs are modified in the host self-DNA, usually by methylation, to prevent the host genome from being targeted (with the exception of type IV R-M systems, which target modified phage DNA while the host genome remains unaltered). R-M systems are classified into four types and are present in more than 74% of prokaryotic genomes. On average, a bacterial genome encodes two R-M systems. DNA modification as a strategy to discriminate between self-DNA and non-self-DNA is not limited to methylation. For example, the dnd defense system modifies the host DNA backbone to include a sulfur group, and the dpd system utilizes a multi-enzyme pathway to modify guanine residues into 7-deazaguanine derivatives in the host DNA. The BREX (bacteriophage exclusion) system and DISARM (defence islands system associated with R-M) also function through methylation of host DNA, although the mechanisms of phage DNA targeting in these systems are still unknown. All of these defence systems constitute part of the innate immunity of bacteria. A large fraction of bacteria and archaea encode CRISPR–Cas, a family of adaptive immune systems that also function through the recognition and degradation of viral nucleic acids. The CRISPR–Cas immune memory is formed through acquisition of short viral-derived DNA sequences that are incorporated as CRISPR ‘spacers’ within the host genome. These sequences are then transcribed and processed into CRISPR RNAs that guide the CRISPR–Cas machinery, through sequence complementarity, to target the viral nucleic acids. CRISPR–Cas systems are diverse, comprising two classes, six types and more than 20 subtypes that differ in the composition of the interference machinery, their mechanisms of targeting and the nucleic acid targeted (that is, DNA or RNA). In most cases, both spacer acquisition and interference necessitate the occurrence of a short sequence motif named PAM (protospacer adjacent motif) next to the sequence matched by the spacer in the targeted molecule. Operons that include prokaryotic argonautes have also been hypothesized to provide defense. Present in 9% and 32% of bacterial and archaeal genomes, respectively, their frequent localization in defense islands (regions in microbial genomes in which defense systems are concentrated) as well as their protective activity against plasmids suggest that they are involved in antiviral defense. Another common strategy of defense against phages is Abi. Abi systems allow the bacterial cell, once infected, to kill itself or to arrest its metabolism before the phage reproductive cycle is completed, thus preventing the phage from spreading and killing the surrounding bacterial community. Abi systems have been detected in a wide variety of microorganisms but, given their high diversity, it is challenging to assess their abundance in nature. These systems are usually triggered by a specific component that could be a phage protein, a nucleic acid or a cellular state caused by phage infection. For example, the Escherichia coli Lit Abi is activated upon sensing a unique substrate formed by the Gol peptide of phage T4 when bound to the ribosomal elongation factor EF-Tu. Once active, the Lit protein cleaves EF-Tu, thus inhibiting translation and ultimately killing the cell.

Box 1: Antiviral defense systems tend to cluster on bacterial and archaeal chromosomes in regions denoted as defense islands. Defense islands typically comprise diverse defense systems. see the figure for examples of defense islands within selected bacterial and archaeal genomes (different colors represent different defense systems). Beige represents genes of non-defense functions or of unknown function. Defense systems are also enriched with genes typical of mobile genetic elements such as transposases, recombinases, and conjugation genes. Some defense islands were predicted to encode more than 100 defense genes. The origin and the mechanism of formation of defense islands are currently unknown but could reflect different effects. First, co-localization of defense genes with mobile genes could facilitate the horizontal transfer of multiple defense systems from one bacterium to another in a single transfer event. Alternatively, such islands can be hotspots for integration of horizontally acquired genes, with defense systems clustering in defense islands through the ‘garbage and pile effect’, in which high rates of acquisition and loss are not strongly deleterious. in addition, such co-localization of defense genes could reflect functional links between the defense systems, including possible co-regulation or positive epistasis. the phenomenon of defense islands in bacterial genomes allows the prediction of novel defense systems through a ‘guilt by association approach. in this approach, protein families with unknown functions that are enriched in defense islands can be predicted to constitute new defense systems. This methodology has led to the discovery of individual defense systems such as BreX (bacteriophage exclusion) or DisarM (defense islands system associated with r-M), and its application in a systematic manner recently revealed nine new antiviral systems that are widespread in bacteria and archaea. For r-M and CrisPr–Cas systems, the type is indicated in parentheses.
Another example is PrrC in E. coli, which cleaves bacterial tRNALys molecules when it senses that the phage has suppressed bacterial R-M systems. In Lactococcus spp., many Abi genes (around 20) have been described: for example, AbiZ accelerates lysis before phage assembly whereas AbiB leads to non-specific degradation of mRNAs. In Staphylococcus spp., the serine threonine kinase STK2 protein is activated when exposed to the phage protein PacK, leading to phosphorylation of proteins involved in multiple cellular pathways and eventual cell death. Toxin–antitoxin systems, representing a large family of two-gene modules each comprising a toxin and an immunity component, were also shown to execute Abi in some cases, although their general role in defense against phages is still disputed. A newly discovered system called CBASS (cyclic oligonucleotide-based anti-phage signalling system) employs a specific form of Abi34. This system, appearing in more than 10% of sequenced bacterial genomes, relies on cyclic oligonucleotide signalling to provide defense against phages. Sensing of phage infection leads to the production of a cyclic oligonucleotide, for example cyclic GMP–AMP, which activates a downstream effector that causes cell death. The CBASS system is considered the prokaryotic ancestor of the cGAS–STING antiviral pathway in animals, which similarly relies on cyclic GMP–AMP signalling. Recent studies have revealed the existence of many additional families of antiviral defense systems in bacteria and archaea. An effort to map microbial defense islands (Box 1) has resulted in the discovery of nine new defense systems that are widespread in bacterial and archaeal genomes. These systems were named after protective deities from world mythologies including Hachiman, Thoeris, Zorya, Gabija and Shedu, and their molecular mechanisms of action are yet to be deciphered. Finally, species of Streptomyces produce small molecules called doxorubicin and daunorubicin that act as DNA intercalants, and were recently shown to specifically block phage DNA replication but not the replication of bacterial DNA4 .
A need for multiple defences
Analysis of sequenced prokaryotic genomes demonstrates that they can concomitantly harbour multiple different defense systems. As shown in Fig. 2, a single strain can encode diverse defence strategies including Abi, R-M and CRISPR–Cas. Many bacteria and archaea encode multiple defence systems of the same kind: for example, Helicobacter pylori F30 encodes three type I R-M systems, 11 type II R-M systems, one type III R-M system and one type IV R-M system15. In total, it was estimated that up to 10% of some prokaryotic genomes is dedicated to defence systems8 . These observations raise a basic question — what is the benefit for a single microorganism to encode so many different lines of defence?

Fig. 2 | closely related bacterial strains encode diverse defense systems. Each line represents a different strain of either Escherichia coli (part a) or Pseudomonas aeruginosa (part b). Each column corresponds to a different defense system (color indicates the presence of a defense system). CRISPR–Cas systems were detected using CRISPRCasFinder, restriction-modification (R-M) systems using HHsearch with HMM profiles, BREX (bacteriophage exclusion) by the presence of the pglZ gene, DISARM (defense islands system associated with R-M) and CBASS (cyclic oligonucleotide-based anti-phage signalling system). For R-M and CRISPR–Cas systems, the type or subtype is indicated in parentheses. R-M, restriction-modification.
One obvious answer is that some defence systems can protect only from a specific type of virus. For example, the GmrSD type IV R-M system only targets phages such as T4, whose genomes are modified to include glucosylated hydroxymethylcytosine. Cas9, on the other hand, cannot cleave the DNA of phage T4 owing to its heavily modified cytosine residues. The Thoeris defence system seems to protect only against phages from the Myoviridae family . Therefore, for a microorganism to be protected against a wide variety of viruses, it should encode a broad defence arsenal that can overcome the multiple types of viruses that can infect it. There are benefits for a microorganism to encode multiple defense systems, even if these systems overlap in the range of viruses that they target. This is because phages can develop resistance to defense. First, phage genomes can evolve to eliminate specific sequences such as motifs targeted by restriction enzymes or PAM sequences that are essential for CRISPR–Cas defense. Second, phages often encode anti-defense proteins, including anti-CRISPR and anti-restriction proteins. These proteins are either injected into the cell together with the viral DNA or expressed early upon infection, and inhibit the defense systems. Anti-CRISPRs are typically short proteins that bind the CRISPR–Cas complex and prevent it from working properly. Recent discoveries report on anti-CRISPRs working as enzymes that can cleave the CRISPR RNA or add an acetyl group to a PAM-sensing residue in the Cas effector. Similarly, anti-restriction proteins inhibit restriction enzymes: for example, the T4 IPI (internal protein I) inhibits type IV R-M systems, whereas the DarA and DarB proteins of phage P1 bind the restriction sites on the phage genome and mask them from cleavage by the type I R-M system of E. coli. Faced with viruses that encode counter-defense mechanisms, bacteria and archaea cannot rely on a single defense system and thus need to present several lines of defense as a bet-hedging strategy of survival.
Gain and loss of defense systems
Owing to the selective advantage that defense systems provide, they are frequently gained by bacteria and archaea through horizontal gene transfer (HGT). Multiple studies based on phylogenetic analyses and comparative genomics have confirmed the high rate of transfer of defense systems. For example, only ∼4% of R-M systems are found in the core genomes of prokaryotic species, suggesting recent transfer events. In another example, an analysis of phylogenetic trees of Cas proteins and CRISPR repeats showed weak consistency with the species tree, demonstrating the dominance of horizontal transfer for the spread of CRISPR–Cas loci. Both CRISPR–Cas and R-M systems have been detected on mobile genetic elements, such as plasmids, transposons and phages, partially explaining their mode of HGT. In addition, genomic analyses have shown that defense systems tend to be concentrated in ‘defense islands’ — regions of the host chromosome that are also enriched with mobile elements presumably responsible for the genetic mobilization of the islands (Box 1). Given their selective advantage in the arms race against viruses, one might expect that defense systems, once acquired (either through direct evolution or via HGT), would accumulate in prokaryotic genomes and be selected for. Surprisingly, this is not the case as defense systems are known to be frequently lost from microbial genomes over short evolutionary time scales, suggesting that they can impose selective disadvantages in the absence of infection pressure. A major drawback of defense systems is autoimmunity: CRISPR–Cas, for example, can make mistakes in the process of spacer acquisition and acquire spacers from the chromosome instead of from the invading element. This directs the CRISPR–Cas interference machinery to attack the chromosome, resulting in cell death or in survival through pseudogenization and eventual deletion of the CRISPR–Cas locus. Similarly, R-M systems can also rarely target the chromosome, cleaving self-DNA at a low but measurable rate and inflicting a fitness cost. Unwanted activity of Abi systems can also lead to dormancy or cell death. In addition to autoimmunity, defense systems can impose an energy burden on the cell: some R-M systems require the hydrolysis of one ATP molecule per base pair for translocation of the restriction enzyme along the DNA. As a result of these fitness costs, there is a selective pressure for bacteria to get rid of defense systems under conditions when there is no selection pressure exerted by phages. Indeed, competition studies between strains encoding defense systems, such as CRISPR–Cas or Lit Abi, and cognate defense-lacking strains have demonstrated the existence of a fitness cost in the absence of phage infection. An experimental study in Staphylococcus epidermidis showed that the loss of CRISPR–Cas systems by large deletions have little or no fitness cost. Another study demonstrated that the inactivation of CRISPR–Cas systems in Streptococcus pneumoniae is even advantageous under specific conditions. The frequent gain and loss of defense systems over short time scales lead to a highly variable pattern of presence and absence of systems in microbial genomes. Even in closely related strains with otherwise similar genomes, the composition of defense systems can drastically vary, as demonstrated in Fig. 2. Defence systems appear to be in a state of constant genetic flux, constituting the second most dynamic class of genes after mobile genetic elements in terms of rates of gain and loss in microbial genomes.
Pan-immunity as a shared resource
Given the fitness costs inflicted by antiviral systems, it is probable that no single bacterial or archaeal strain can encode, in the long term, all possible defense systems without suffering serious competitive disadvantages. On the other hand, access to a diverse set of defense mechanisms is essential in order to combat the enormous genetic and functional diversity of viruses. We propose that these seemingly contradictory requirements can be reconciled when considering the available arsenal of immune systems as a resource shared by a population of bacteria or archaea rather than by individual cells. In the example shown in Fig. 2, none of the strains encode all defense systems. However, if these strains are mixed as part of a population, the pan-genome of this population would encode an ‘immune potential’ that encompasses all of the depicted systems. As these systems can be readily available by HGT, given the high rate of HGT in defense systems, the population in effect harbors an accessible reservoir of immune systems that can be acquired by population members. When the population is subjected to infection, this diversity ensures that at least some population members would encode the appropriate defense system, and these members would survive and form the basis for the perpetuation of the population (Fig. 3).

The pan-immune system model.
Closely related strains of microorganisms within a population encode a diverse set of antiviral systems constituting the pan-immune system defense.
a | Maintenance of diversity of the pan-immune system. Bacteriophage (phage) infection results in bacterial selection for those encoding a specific defense system (red) that can overcome that phage (part a, stage 1). In the absence of phage pressure over a period of time, members of the population can acquire a diverse set of defense systems, through horizontal gene transfer (HGT) (part a, stage 2), whereas some cells lose defense systems owing to their selective cost (part a, stage 3). The cycle continues, resulting in a population defense that together constitutes the immune potential of the population.
b | Dynamic changes to the pan-immune system composition. Phage infection results in selection for members encoding a specific defense system (yellow) that can overcome that phage (part b, stage 1). In some cases, this can result in the loss of immune system defense (green). Conversely, new systems can be introduced into the population through HGT from a more distantly related strain, migration of a new member in the population or emergence of a new system through mutation or exaptation (part b, stage 2). Red, green, yellow and blue genes represent different types of defense systems.
We thus hypothesize that some of the selection for defense systems occurs at the group level. In a sense, this pan-immune system model aligns well with previous observations and mathematical models of distributed immunity that specifically focused on CRISPR–Cas systems. Studies on CRISPR–Cas have shown that spacer diversity in the population is essential to overcoming phage infections. In co-evolution experiments between Pseudomonas aeruginosa and Streptococcus thermophilus and their respective phages, bacterial populations in which different strains encoded different sets of spacers overcame phage infection and resulted in phage extinction, whereas populations comprising homogeneous sets of spacers allowed phage propagation. Protection of spacer-diverse populations occurred because no single phage could accumulate enough mutations to overcome the diversity of spacers encoded by the population as a whole. In the context of CRISPR–Cas, mathematical models that explored the parameters leading to the emergence of a distributed immunity predicted two key parameters: the cost of generating a new allele (in this case, a new spacer) should be small; and the fitness constraints of evolving escape mutations for phages is enhanced by the fact that an escape mutant will be resistant only to one allele (one spacer in the case of the CRISPR model). Beyond the specific case of CRISPR–Cas, the same conditions also fit the broader context of the microbial pan-immune system model, which can be viewed as satisfying the two parameters mentioned above: given the high rate of HGT of defense systems (which can be considered the acquisition of alleles of defense), the cost of acquiring a new allele via HGT is expected to be relatively small; and due to the diversity of molecular mechanisms among different defense systems, the emergence of one phage mutation that allows escape from a specific defense system is not expected to abolish defense by other systems. As group selection occurs within closely related kin, we expect the pan-immune system model to be mainly relevant among populations of similar, related strains that differ in their defense content, thus allowing for selection at the group level.
Implications for counter-defence
It is well documented that individual phages have well-defined host ranges, such that they can infect some, but rarely all, strains of the same species. This is often attributed to the diversity of surface molecules among the infected microbial strains, as these are used by phages as specific receptors. However, given the diversity of defense systems observed in different strains of the same species, it is clear that the host range of any given phage would depend on its ability to overcome multiple defence systems. This predicts that phages need to encode many different counter-defense mechanisms in order to have a broad host range. This prediction may help reconcile the puzzle of dispensable genes in phage genomes. As phage genomes are under strong selection, one might expect that most of their genes are essential. However, serial mutational analyses showed that as much as 79% of genes in phage T4 and 63% of genes in phage T7 are not essential for successful infection of the E. coli laboratory strain. We predict that many of these genes will turn out to encode anti-defense proteins that target defense systems not present in the E. coli strain used in these studies. We would therefore expect that the set of anti-defense genes cumulatively encoded by strains of a phage species should mirror the set of defense systems encoded by its host pan-genome. 1
Evolution of Immune Systems From Viruses and Transposable Elements
Felix Broecker (2019): Cellular organisms have co-evolved with various mobile genetic elements (MGEs), including transposable elements (TEs), retroelements, and viruses, many of which can integrate into the host DNA. MGEs constitute ∼50% of mammalian genomes, >70% of some plant genomes, and up to 30% of bacterial genomes. 2

FIGURE 1. Cartoons depicting various defense systems.
The systems are color-coded based on the level of support in green (experimental evidence available), magenta (bioinformatic evidence available), and orange (speculative with some supporting evidence). DNA/RNA cleaving is indicated with scissors. RNA is depicted as wavy lines or with secondary hairpin-loop structures. A ribozyme cleaving another ribozyme is a hypothetical early immune system that does not require DNA or proteins. The ribozyme may be part of a viroid-like selfish RNA. Restriction-modification systems distinguish between foreign and self DNA by methylating target sequences of restriction endonucleases. Prophages can mediate superinfection exclusion, exemplified by the expression of the Tip protein that reduces bacterial surface expression of type IV pili required for infection by various phages. CRISPR-Cas acts by incorporating small genomic fragments from phages into CRISPR arrays in the prokaryotic genome. The transcribed spacers are then used by another Cas member to cleave sequence-homologous phage genomes. PIWI-associated RNAs (piRNAs) are small RNAs complementary to transposable elements (TEs) that are encoded in piRNA clusters. piRNAs associate with a PIWI nuclease to cleave complementary TE transcripts. RNA interference (RNAi) is initiated by dsRNA which is fragmented by Dicer to siRNAs. These siRNAs are loaded into the RNA-induced silencing complex to cleave complementary RNAs using Ago nucleases. A variation of RNAi is the endo-siRNA pathway, in which dsRNA is generated from TEs that are transcribed in both orientations, for instance, if the TE is located in an intron in opposite orientation to the encompassing gene. Endogenous retroviruses (ERVs) can mediate the restriction of ERVs and exogenous retroviruses through various mechanisms, including receptor blockade by captured Env proteins, Gag-mediated restriction, and antisense RNA mechanisms. The interferon system recognizes dsRNA or other pathogen-associated molecular patterns, which leads to the upregulation of antiviral interferon-stimulated genes (ISGs). The antibody system involves diversification through light and heavy chain recombination, which is mediated by the Rag recombinases. This enables the detection of diverse pathogens. Note that the references provided in the figure are not comprehensive. Please refer to the text for more details and additional references.
1. Aude Bernheim: The pan-immune system of bacteria: antiviral defence as a community resource 06 November 2019
2. Felix Broecker: Evolution of Immune Systems From Viruses and Transposable Elements 29 January 2019

