Prebiotic origin of DNA replication
https://reasonandscience.catsboard.com/t3119-prebiotic-origin-of-dna-replication
Reproduction is important for the survival of all living things. Without a mechanism for reproduction, life would come to an end. 1
Bacteria reproduce by binary fission. Binary fission begins when the DNA of the bacterium divides into two (replicates). 2
DNA replication, and its mind boggling nano technology that defies naturalistic explanations
https://reasonandscience.catsboard.com/t1849-dna-replication-of-prokaryotes
The Argument of the Original Replicator
In prokaryotic cells, DNA replication involves more than thirty specialized proteins to perform tasks necessary for building and accurately copying the genetic molecule.
Each of these proteins is essential and required for the proper replicating process. Not a single one of these proteins can be missing, otherwise, the whole process breaks down, and is unable to perform its task correctly. DNA repair mechanisms must also be in place, fully functional, and working properly, otherwise, the mutation rate will be too high, and the cell dies. 18
The individual parts and proteins require by themselves complex assembly proteins to be built.
The individual parts, assembly proteins, and proteins individually would have no function on their own. They have only function interconnected in the working whole.
The individual parts must be readily available on the construction site of the DNA replication complex, being correctly interlocked, interlinked, and have the right interface compatibility to be able to interact correctly together. All this requires information and meta-information ( information that directs the expression of the genomic information for construction of the individual proteins, and correct timing of expression, and as well the information of the correct assembly sequence. )
Evolution is not a capable driving force to make the DNA replicating complex, because evolution depends on cell replication through the very own mechanism we try to explain. It takes proteins to make DNA replication happen. But it takes the DNA replication process to make proteins. That’s a catch 22 situation.
DNA replication requires coded, complex, specified information and meta-information, and the DNA replication process is irreducibly complex.
Therefore, DNA replication is best explained through design.
Tan, Change; Stadler, Rob. The Stairway To Life:
Combining the nucleotides
The natural replication process of DNA for E. coli has an error rate of approximately 1 in 1,000,000,000. It is not only incredibly accurate, but it is also incredibly fast (about one thousand nucleotides per second in E. coli). If DNA was scaled up to be one meter in diameter, the protein-based molecular machinery that replicates DNA “would move at approximately 600 km/hr (375 mph), and the replication machinery would be about the size of a FedEx delivery truck. Replicating the E. coli genome would be a 40 min, 400 km (250 mile) trip for two such machines, which would, on average make an error only once every 170 km (106 miles)”
C.Venter Design and synthesis of a minimal bacterial genome 2016
https://sci-hub.ren/10.1126/science.aad6253
The replication of genomic information and its coordinated distribution into segregated membranebound cellular compartments are hallmarks of extant living systems that are commonly considered to be among the attributes that define cellular life. The minimal requirements for this process are not known, but evidence from disparate fields of study suggests that mechanisms mechanisms far simpler than the complex division apparatus in most eubacteria may be sufficient.
DNA replication 16 Genes

Determination of the Core of a Minimal Bacterial Gene Set Sept. 2004
https://mmbr.asm.org/content/mmbr/68/3/518.full.pdf
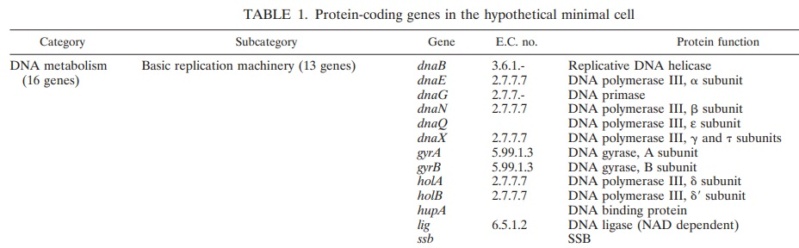
DNA metabolism. (i) Basic replication machinery.
One of the most important housekeeping functions in all living cells is DNA replication, a multistage reaction with four basic steps.
First, a replication origin sequence within the genome is recognized by protein components.
Second, initiation of replication occurs through the recruitment of replisomal proteins at the origin.
Third, the general replication reaction duplicates both strands of DNA.
Fourth, replication terminates and the two daughter chromosomes are separated.
The specific mechanism for replication initiation depends both on the structure of the replication origin and on the nature of the initiation protein. Several mechanisms have been described in bacteria, but the analysis of the genome of B. floridanus, the endosymbiotic bacteria of carpenter ants, revealed that it does not encode any of the proposed initiation proteins (DnaA, RecA, and PriA). Another recruitment protein (DnaC in E. coli, DnaI in B. subtilis) is also required to form a complex with the helicase DnaB (EC 3.6.1.-) to prevent the indiscriminate binding of the helicase to single-stranded DNA, transferring it only in the correct DNA location which is not coated by Single-Stranded DNA Binding protein. (SSB). However, DnaC is absent in B. floridanus and W. glossinidia (endosymbiont of tsetse flies), indicating that it is also dispensable in these bacteria. Hence, it can be assumed that DNA replication can take place without the need for these initiation and recruiting proteins under some conditions, which might be the case for small genomes, or that there are other, as yet unidentified proteins that perform such functions in these reduced genomes. The recruitment and loading of helicase at the replication origin generally requires the presence of a histonelike protein for the destabilization of the duplex DNA near the origin of replication. At least one histone-like protein is also encoded by all five analyzed endosymbiont genomes, although none of these proteins is conserved in all of them. In M. genitalium, MG353 appears to encode a nucleoid DNA binding protein similar to hupA. Since this gene is also present in all endosymbionts analyzed except B. floridanus, we decided to include this histone-like protein-coding gene in our minimal set. To begin DNA replication, the helicase DnaB (EC 3.6.1.-) attracts the primase DnaG (EC 2.7.7.-) to the replication fork, both of them present in all bacteria with reduced genomes analyzed in this study. Once the DNA is melted and primed, the general DNA replication begins, with the action of DNA polymerase III (EC 2.7.7.7). Genes encoding the subunits of DNA polymerase III, all subunits that are present both in gram-negative and gram-positive bacteria, are also conserved. It should be noticed that in gram-positive bacteria there are two genes encoding the subunits of this holoenzyme, both of them essential in B. subtilis, but one of them (dnaE) has been proven to be dispensable in M. genitalium, while polC encodes both the and ε subunits of DNA polymerase III. Two more proteins required at this stage, gyrase (EC 5.99.1.2, encoded by gyrA and gyrB) and ligase (EC 6.5.1.2, encoded by lig), are also conserved, thus allowing the completion of the third stage of DNA replication. Although lig was previously annotated as a pseudogene in B. aphidicola BSg due to the absence of the C-terminal protein end (80), it has been reannotated as a functional gene encoding the catalytic domain of the protein but with the absence of the BRCT domain (28). On the other hand, the gene encoding the protein necessary to stop replication by inhibiting helicase translocation (tus in E. coli, rtp in B. subtilis) cannot be found in the reduced genomes under study. Several topoisomerases that are present in M. genitalium and essential in B. subtilis and E. coli to allow independent segregation of the two daughter chromosomes (EC 5.99.1.2, encoded by topA, and EC 5.99.1.-, encoded by parC and parE) must also be considered dispensable, since parC and parE have been lost in all endosymbionts analyzed, while topA is only present in two strains of B. aphidicola. It is possible that the above-mentioned GyrA/B, which is present in all these bacteria, performs all the topoisomerase functions required for DNA replication and segregation, since it has been suggested that type IA (TopA) and type II (GyrA/B and ParD/E) topoisomerases may use comparable mechanisms to bind and cleave DNA, based on similarities in several structural elements (6). Other proteins involved in chromosome condensation that are essential in B. subtilis (SMC, ScpA, and ScpB) are not conserved either, and they have not been included in the minimal set. SMC proteins are conserved in most (but not all) prokaryotes, and E. coli mutants lacking the encoding gene are viable.
Helicase
Helicases are astonishing motor proteins which rotational speed is up to 10,000 rotations per minute, and are life essential.
How Many Genes Can Make a Cell: The Minimal-Gene-Set Concept
https://www.ncbi.nlm.nih.gov/books/NBK2227/
We propose a minimal gene set composed of 206 genes. Such a gene set will be able to sustain the main vital functions of a hypothetical simplest bacterial cell with the following features.
(i) A virtually complete DNA replication machinery, composed of one nucleoid DNA binding protein, SSB, DNA helicase, primase, gyrase, polymerase III, and ligase. No initiation and recruiting proteins seem to be essential, and the DNA gyrase is the only topoisomerase included, which should perform both replication and chromosome segregation functions.
Helicase are a class of enzymes vital to all living organisms. Their main function is to unpackage an organism's genes, and they are essential for dna replication, and evolution to be able to occur. They require 1000 left-handed amino acids in the right specified sequence. Each of the 1000 amino acids must be the right amongst 20 to chose from. How did they emerge by natural processes? The chance to get them by random chemical reactions is 1 in 20^1000 or 1 in 10^1300..... there are 10^80 atoms in the universe.
The odds are in reality much greater. There exist hundreds of different amino acids, which supposedly were extant on the early earth. Amongst these, an unknown selection process would have had to select the 20 amino acids used in life, select only left-handed ones from a mix of left and right handed ones.
But the problems do not stop there.
Cairns-Smith, the Genetic Takeover, page 59:
For one overall reaction, making one peptide bond, there about 90 distinct operations are required. If you were to consider in more detail a process such as the purification of an intermediate you would find many subsidiary operations — washings, pH changes and so on.
1. The synthesis of proteins and nucleic acids from small molecule precursors, and the formation of amide bonds without the assistance of enzymes represents one of the most difficult challenges to the model of pre-vital ( chemical) evolution, and for theories of the orgin of life.
2. The best one can hope for from such a scenario is a racemic polymer of proteinous and non-proteinous amino acids with no relevance to living systems.
3. Polymerization is a reaction in which water is a product. Thus it will only be favoured in the absence of water. The presence of precursors in an ocean of water favours depolymerization of any molecules that might be formed.
4. Even if there were billions of simultaneous trials as the billions of building block molecules interacted in the oceans, or on the thousands of kilometers of shorelines that could provide catalytic surfaces or templates, even if, as is claimed, there was no oxygen in the prebiotic earth, then there would be no protection from UV light, which would destroy and disintegrate prebiotic organic compounds. Secondly, even if there would be a sequence, producing a functional folding protein, by itself, if not inserted in a functional way in the cell, it would absolutely no function. It would just lay around, and then soon disintegrate. Furthermore, in modern cells proteins are tagged and transported on molecular highways to their precise destination, where they are utilized. Obviously, all this was not extant on the early earth.
5. To form a chain, it is necessary to react bifunctional monomers, that is, molecules with two functional groups so they combine with two others. If a unifunctional monomer (with only one functional group) reacts with the end of the chain, the chain can grow no further at this end. If only a small fraction of unifunctional molecules were present, long polymers could not form. But all ‘prebiotic simulation’ experiments produce at least three times more unifunctional molecules than bifunctional molecules.
1. https://www.msnucleus.org/membership/html/k-6/lc/organ/5/lco5_4a.html#:~:text=Reproduction%20is%20important%20for%20the,would%20come%20to%20an%20end.&text=Some%20single-celled%20organisms%20reproduce,half%20producing%20two%20daughter%20cells.
2. https://microbiologysociety.org/why-microbiology-matters/what-is-microbiology/bacteria.html#:~:text=Bacteria%20reproduce%20by%20binary%20fission,into%20two%20identical%20daughter%20cells.&text=The%20bacterial%20cell%20then%20elongates,clone%20of%20the%20parent%20cell.
https://reasonandscience.catsboard.com/t3119-prebiotic-origin-of-dna-replication
Reproduction is important for the survival of all living things. Without a mechanism for reproduction, life would come to an end. 1
Bacteria reproduce by binary fission. Binary fission begins when the DNA of the bacterium divides into two (replicates). 2
DNA replication, and its mind boggling nano technology that defies naturalistic explanations
https://reasonandscience.catsboard.com/t1849-dna-replication-of-prokaryotes
The Argument of the Original Replicator
In prokaryotic cells, DNA replication involves more than thirty specialized proteins to perform tasks necessary for building and accurately copying the genetic molecule.
Each of these proteins is essential and required for the proper replicating process. Not a single one of these proteins can be missing, otherwise, the whole process breaks down, and is unable to perform its task correctly. DNA repair mechanisms must also be in place, fully functional, and working properly, otherwise, the mutation rate will be too high, and the cell dies. 18
The individual parts and proteins require by themselves complex assembly proteins to be built.
The individual parts, assembly proteins, and proteins individually would have no function on their own. They have only function interconnected in the working whole.
The individual parts must be readily available on the construction site of the DNA replication complex, being correctly interlocked, interlinked, and have the right interface compatibility to be able to interact correctly together. All this requires information and meta-information ( information that directs the expression of the genomic information for construction of the individual proteins, and correct timing of expression, and as well the information of the correct assembly sequence. )
Evolution is not a capable driving force to make the DNA replicating complex, because evolution depends on cell replication through the very own mechanism we try to explain. It takes proteins to make DNA replication happen. But it takes the DNA replication process to make proteins. That’s a catch 22 situation.
DNA replication requires coded, complex, specified information and meta-information, and the DNA replication process is irreducibly complex.
Therefore, DNA replication is best explained through design.
Tan, Change; Stadler, Rob. The Stairway To Life:
Combining the nucleotides
The natural replication process of DNA for E. coli has an error rate of approximately 1 in 1,000,000,000. It is not only incredibly accurate, but it is also incredibly fast (about one thousand nucleotides per second in E. coli). If DNA was scaled up to be one meter in diameter, the protein-based molecular machinery that replicates DNA “would move at approximately 600 km/hr (375 mph), and the replication machinery would be about the size of a FedEx delivery truck. Replicating the E. coli genome would be a 40 min, 400 km (250 mile) trip for two such machines, which would, on average make an error only once every 170 km (106 miles)”
C.Venter Design and synthesis of a minimal bacterial genome 2016
https://sci-hub.ren/10.1126/science.aad6253
The replication of genomic information and its coordinated distribution into segregated membranebound cellular compartments are hallmarks of extant living systems that are commonly considered to be among the attributes that define cellular life. The minimal requirements for this process are not known, but evidence from disparate fields of study suggests that mechanisms mechanisms far simpler than the complex division apparatus in most eubacteria may be sufficient.
DNA replication 16 Genes

Determination of the Core of a Minimal Bacterial Gene Set Sept. 2004
https://mmbr.asm.org/content/mmbr/68/3/518.full.pdf

DNA metabolism. (i) Basic replication machinery.
One of the most important housekeeping functions in all living cells is DNA replication, a multistage reaction with four basic steps.
First, a replication origin sequence within the genome is recognized by protein components.
Second, initiation of replication occurs through the recruitment of replisomal proteins at the origin.
Third, the general replication reaction duplicates both strands of DNA.
Fourth, replication terminates and the two daughter chromosomes are separated.
The specific mechanism for replication initiation depends both on the structure of the replication origin and on the nature of the initiation protein. Several mechanisms have been described in bacteria, but the analysis of the genome of B. floridanus, the endosymbiotic bacteria of carpenter ants, revealed that it does not encode any of the proposed initiation proteins (DnaA, RecA, and PriA). Another recruitment protein (DnaC in E. coli, DnaI in B. subtilis) is also required to form a complex with the helicase DnaB (EC 3.6.1.-) to prevent the indiscriminate binding of the helicase to single-stranded DNA, transferring it only in the correct DNA location which is not coated by Single-Stranded DNA Binding protein. (SSB). However, DnaC is absent in B. floridanus and W. glossinidia (endosymbiont of tsetse flies), indicating that it is also dispensable in these bacteria. Hence, it can be assumed that DNA replication can take place without the need for these initiation and recruiting proteins under some conditions, which might be the case for small genomes, or that there are other, as yet unidentified proteins that perform such functions in these reduced genomes. The recruitment and loading of helicase at the replication origin generally requires the presence of a histonelike protein for the destabilization of the duplex DNA near the origin of replication. At least one histone-like protein is also encoded by all five analyzed endosymbiont genomes, although none of these proteins is conserved in all of them. In M. genitalium, MG353 appears to encode a nucleoid DNA binding protein similar to hupA. Since this gene is also present in all endosymbionts analyzed except B. floridanus, we decided to include this histone-like protein-coding gene in our minimal set. To begin DNA replication, the helicase DnaB (EC 3.6.1.-) attracts the primase DnaG (EC 2.7.7.-) to the replication fork, both of them present in all bacteria with reduced genomes analyzed in this study. Once the DNA is melted and primed, the general DNA replication begins, with the action of DNA polymerase III (EC 2.7.7.7). Genes encoding the subunits of DNA polymerase III, all subunits that are present both in gram-negative and gram-positive bacteria, are also conserved. It should be noticed that in gram-positive bacteria there are two genes encoding the subunits of this holoenzyme, both of them essential in B. subtilis, but one of them (dnaE) has been proven to be dispensable in M. genitalium, while polC encodes both the and ε subunits of DNA polymerase III. Two more proteins required at this stage, gyrase (EC 5.99.1.2, encoded by gyrA and gyrB) and ligase (EC 6.5.1.2, encoded by lig), are also conserved, thus allowing the completion of the third stage of DNA replication. Although lig was previously annotated as a pseudogene in B. aphidicola BSg due to the absence of the C-terminal protein end (80), it has been reannotated as a functional gene encoding the catalytic domain of the protein but with the absence of the BRCT domain (28). On the other hand, the gene encoding the protein necessary to stop replication by inhibiting helicase translocation (tus in E. coli, rtp in B. subtilis) cannot be found in the reduced genomes under study. Several topoisomerases that are present in M. genitalium and essential in B. subtilis and E. coli to allow independent segregation of the two daughter chromosomes (EC 5.99.1.2, encoded by topA, and EC 5.99.1.-, encoded by parC and parE) must also be considered dispensable, since parC and parE have been lost in all endosymbionts analyzed, while topA is only present in two strains of B. aphidicola. It is possible that the above-mentioned GyrA/B, which is present in all these bacteria, performs all the topoisomerase functions required for DNA replication and segregation, since it has been suggested that type IA (TopA) and type II (GyrA/B and ParD/E) topoisomerases may use comparable mechanisms to bind and cleave DNA, based on similarities in several structural elements (6). Other proteins involved in chromosome condensation that are essential in B. subtilis (SMC, ScpA, and ScpB) are not conserved either, and they have not been included in the minimal set. SMC proteins are conserved in most (but not all) prokaryotes, and E. coli mutants lacking the encoding gene are viable.
Helicase
Helicases are astonishing motor proteins which rotational speed is up to 10,000 rotations per minute, and are life essential.
How Many Genes Can Make a Cell: The Minimal-Gene-Set Concept
https://www.ncbi.nlm.nih.gov/books/NBK2227/
We propose a minimal gene set composed of 206 genes. Such a gene set will be able to sustain the main vital functions of a hypothetical simplest bacterial cell with the following features.
(i) A virtually complete DNA replication machinery, composed of one nucleoid DNA binding protein, SSB, DNA helicase, primase, gyrase, polymerase III, and ligase. No initiation and recruiting proteins seem to be essential, and the DNA gyrase is the only topoisomerase included, which should perform both replication and chromosome segregation functions.
Helicase are a class of enzymes vital to all living organisms. Their main function is to unpackage an organism's genes, and they are essential for dna replication, and evolution to be able to occur. They require 1000 left-handed amino acids in the right specified sequence. Each of the 1000 amino acids must be the right amongst 20 to chose from. How did they emerge by natural processes? The chance to get them by random chemical reactions is 1 in 20^1000 or 1 in 10^1300..... there are 10^80 atoms in the universe.
The odds are in reality much greater. There exist hundreds of different amino acids, which supposedly were extant on the early earth. Amongst these, an unknown selection process would have had to select the 20 amino acids used in life, select only left-handed ones from a mix of left and right handed ones.
But the problems do not stop there.
Cairns-Smith, the Genetic Takeover, page 59:
For one overall reaction, making one peptide bond, there about 90 distinct operations are required. If you were to consider in more detail a process such as the purification of an intermediate you would find many subsidiary operations — washings, pH changes and so on.
1. The synthesis of proteins and nucleic acids from small molecule precursors, and the formation of amide bonds without the assistance of enzymes represents one of the most difficult challenges to the model of pre-vital ( chemical) evolution, and for theories of the orgin of life.
2. The best one can hope for from such a scenario is a racemic polymer of proteinous and non-proteinous amino acids with no relevance to living systems.
3. Polymerization is a reaction in which water is a product. Thus it will only be favoured in the absence of water. The presence of precursors in an ocean of water favours depolymerization of any molecules that might be formed.
4. Even if there were billions of simultaneous trials as the billions of building block molecules interacted in the oceans, or on the thousands of kilometers of shorelines that could provide catalytic surfaces or templates, even if, as is claimed, there was no oxygen in the prebiotic earth, then there would be no protection from UV light, which would destroy and disintegrate prebiotic organic compounds. Secondly, even if there would be a sequence, producing a functional folding protein, by itself, if not inserted in a functional way in the cell, it would absolutely no function. It would just lay around, and then soon disintegrate. Furthermore, in modern cells proteins are tagged and transported on molecular highways to their precise destination, where they are utilized. Obviously, all this was not extant on the early earth.
5. To form a chain, it is necessary to react bifunctional monomers, that is, molecules with two functional groups so they combine with two others. If a unifunctional monomer (with only one functional group) reacts with the end of the chain, the chain can grow no further at this end. If only a small fraction of unifunctional molecules were present, long polymers could not form. But all ‘prebiotic simulation’ experiments produce at least three times more unifunctional molecules than bifunctional molecules.
1. https://www.msnucleus.org/membership/html/k-6/lc/organ/5/lco5_4a.html#:~:text=Reproduction%20is%20important%20for%20the,would%20come%20to%20an%20end.&text=Some%20single-celled%20organisms%20reproduce,half%20producing%20two%20daughter%20cells.
2. https://microbiologysociety.org/why-microbiology-matters/what-is-microbiology/bacteria.html#:~:text=Bacteria%20reproduce%20by%20binary%20fission,into%20two%20identical%20daughter%20cells.&text=The%20bacterial%20cell%20then%20elongates,clone%20of%20the%20parent%20cell.
Last edited by Otangelo on Sun Mar 14, 2021 9:10 pm; edited 1 time in total

