Multimodal transcriptional control of pattern formation in embryonic development
Multimodal transcriptional control of pattern formation in embryonic development December 27, 2019 1
Predicting how the gene expression patterns that specify animal body plans arise from interactions between transcription factor proteins and regulatory DNA remains a major challenge in physical biology. While the modulation of transcriptional bursting has been implicated as the primary lever for controlling gene expression, we find that this alone cannot quantitatively recapitulate pattern formation. Instead, we find that the pattern arises through the joint action of 2 regulatory strategies—control of bursting and control of the total duration of transcription—that originate from distinct underlying molecular mechanisms. During embryonic development, tightly choreographed patterns of gene expression—shallow gradients, sharp steps, narrow stripes—specify cell fates. The correct positioning, sharpness, and amplitude of these patterns of cytoplasmic mRNA and protein ensure the reliable determination of animal body plans. Predicting developmental outcomes demands a quantitative understanding of the flow of information along the central dogma: how input transcription factors dictate the output rate of mRNA production, how this rate of mRNA production dictates cytoplasmic patterns of mRNA, and how these mRNA patterns lead to protein patterns that feed back into the gene regulatory network. While the connection between transcription factor concentration and output mRNA production rate has been the subject of active research over the last 3 decades, the connection between this output rate and the resulting cytoplasmic patterns of mRNA has remained largely unexplored. For example, a graded stripe of cytoplasmic mRNA within an embryo could arise as a result of radically different transcriptional dynamics at the single-nucleus level
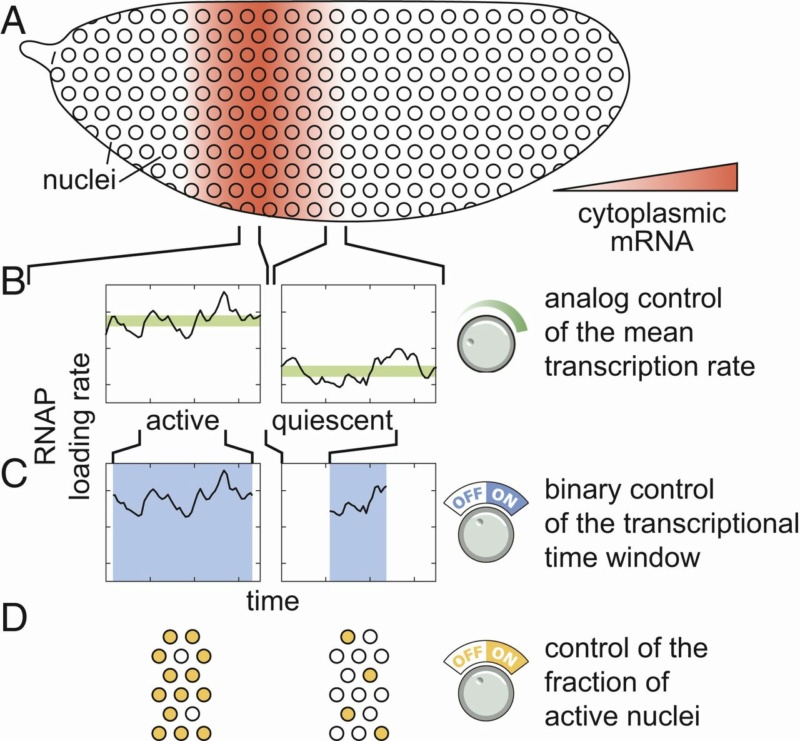
Multiple modes of pattern formation by single-cell transcriptional activity.
(A–D) Cytoplasmic mRNA patterns (A) could arise from transcription factors exerting control over the mean transcription rate (B), the transcriptional time window dictating when a nucleus is transcriptionally active or quiescent (C), or the fraction of active nuclei (D) or some combination thereof.
The detailed cytoplasmic distribution of mRNA that makes these stripes is key to the transmission of spatial information along the gene regulatory network that drives Drosophila development and reinforcing the need to develop models of gene regulation capable of connecting quantitative variations in input transcription factor patterns to graded output rates of transcription. We found that all 3 regulatory strategies outlined in Fig. above quantitatively contribute to the formation of eve stripe 2.
First, a smaller fraction of nuclei become active and engage in transcription in the periphery of the stripe than in the center, although this regulation of the fraction of active nuclei makes only a minor contribution to stripe formation. Second, the rate of mRNA production is significantly elevated in the center of the stripe. This analog control of the transcription rate is insufficient to quantitatively recapitulate the cytoplasmic mRNA stripe pattern. In addition to the control of the rate of mRNA production among nuclei, there is a pronounced regulation of the window of time during which eve loci were engaged in transcription across the stripe, with those in the stripe center expressing for approximately 3 times longer than those in the flanks. While it is widely appreciated that genes are transcriptionally competent for limited windows of time during development, we found that—in the case of eve stripe 2—this binary transcriptionally engaged/disengaged logic is not merely a necessary precondition for pattern formation—it is the primary driver thereof. Thus, we conclude that the regulation of eve stripe 2 is multimodal in nature, with contributions from 3 distinct regulatory strategies (Fig. B–D above). Nonetheless, stripe formation can be quantitatively explained almost entirely through the interplay between 2 distinct control strategies: binary control of the duration of transcriptional engagement. (Fig. C) and control of the mean rate of transcription (Fig. B).
To describe eve stripe 2 transcriptional dynamics, we need to account for both the short, transient ON periods dictated by transcriptional bursts and a longer transcriptional time window that describes the period over which loci engage in this transcriptional bursting. One or more of these bursting parameters are subject to spatially controlled regulation.
In Drosophila development, information encoded in a handful of maternally deposited protein gradients propagates through increasingly complex layers of interacting genes, culminating in the specification of the adult body plan. A priori, there are several distinct regulatory strategies at the single-cell level capable of generating spatially differentiated patterns of cytoplasmic mRNA each with distinct implications for the nature of the underlying molecular processes at play. The average rate of transcription is mainly modulated across the embryo by tuning the frequency of transcriptional bursting. It has remained unclear whether this modulation of the rate of transcription (and thereby mRNA production) is the dominant modality by which input concentrations of transcription factors drive the formation of patterns of gene expression or whether, instead, it is simply the most readily apparent mechanism among multiple distinct control strategies.
While it is widely appreciated that genes are expressed for discrete windows of time over the course of development, in the case of this eve stripe 2 reporter—this binary transcriptionally engaged/quiescent logic is actively regulated by transcription factors to drive pattern formation. Important is the temporal component of transcriptional regulation in specifying developmental outcomes. The limited readout time imposed by short nuclear cycles in early Drosophila development places strict constraints on the kinds of regulatory architecture that could be responsible for driving observed patterns of hunchback gene expression. The pioneer factor Zelda plays a key role in regulating both the timing and probability of transcriptional activation following mitosis. Our work complements these previous observations by exploring yet another aspect of the interplay between timing and transcriptional regulation.
The Drosophila Pioneer Factor Zelda Modulates the Nuclear Microenvironment of a Dorsal Target Enhancer to Potentiate Transcriptional Output April 22, 2019 2
1. https://www.pnas.org/content/117/2/836
2. https://www.sciencedirect.com/science/article/pii/S0960982219303173
Multimodal transcriptional control of pattern formation in embryonic development December 27, 2019 1
Predicting how the gene expression patterns that specify animal body plans arise from interactions between transcription factor proteins and regulatory DNA remains a major challenge in physical biology. While the modulation of transcriptional bursting has been implicated as the primary lever for controlling gene expression, we find that this alone cannot quantitatively recapitulate pattern formation. Instead, we find that the pattern arises through the joint action of 2 regulatory strategies—control of bursting and control of the total duration of transcription—that originate from distinct underlying molecular mechanisms. During embryonic development, tightly choreographed patterns of gene expression—shallow gradients, sharp steps, narrow stripes—specify cell fates. The correct positioning, sharpness, and amplitude of these patterns of cytoplasmic mRNA and protein ensure the reliable determination of animal body plans. Predicting developmental outcomes demands a quantitative understanding of the flow of information along the central dogma: how input transcription factors dictate the output rate of mRNA production, how this rate of mRNA production dictates cytoplasmic patterns of mRNA, and how these mRNA patterns lead to protein patterns that feed back into the gene regulatory network. While the connection between transcription factor concentration and output mRNA production rate has been the subject of active research over the last 3 decades, the connection between this output rate and the resulting cytoplasmic patterns of mRNA has remained largely unexplored. For example, a graded stripe of cytoplasmic mRNA within an embryo could arise as a result of radically different transcriptional dynamics at the single-nucleus level

Multiple modes of pattern formation by single-cell transcriptional activity.
(A–D) Cytoplasmic mRNA patterns (A) could arise from transcription factors exerting control over the mean transcription rate (B), the transcriptional time window dictating when a nucleus is transcriptionally active or quiescent (C), or the fraction of active nuclei (D) or some combination thereof.
The detailed cytoplasmic distribution of mRNA that makes these stripes is key to the transmission of spatial information along the gene regulatory network that drives Drosophila development and reinforcing the need to develop models of gene regulation capable of connecting quantitative variations in input transcription factor patterns to graded output rates of transcription. We found that all 3 regulatory strategies outlined in Fig. above quantitatively contribute to the formation of eve stripe 2.
First, a smaller fraction of nuclei become active and engage in transcription in the periphery of the stripe than in the center, although this regulation of the fraction of active nuclei makes only a minor contribution to stripe formation. Second, the rate of mRNA production is significantly elevated in the center of the stripe. This analog control of the transcription rate is insufficient to quantitatively recapitulate the cytoplasmic mRNA stripe pattern. In addition to the control of the rate of mRNA production among nuclei, there is a pronounced regulation of the window of time during which eve loci were engaged in transcription across the stripe, with those in the stripe center expressing for approximately 3 times longer than those in the flanks. While it is widely appreciated that genes are transcriptionally competent for limited windows of time during development, we found that—in the case of eve stripe 2—this binary transcriptionally engaged/disengaged logic is not merely a necessary precondition for pattern formation—it is the primary driver thereof. Thus, we conclude that the regulation of eve stripe 2 is multimodal in nature, with contributions from 3 distinct regulatory strategies (Fig. B–D above). Nonetheless, stripe formation can be quantitatively explained almost entirely through the interplay between 2 distinct control strategies: binary control of the duration of transcriptional engagement. (Fig. C) and control of the mean rate of transcription (Fig. B).
To describe eve stripe 2 transcriptional dynamics, we need to account for both the short, transient ON periods dictated by transcriptional bursts and a longer transcriptional time window that describes the period over which loci engage in this transcriptional bursting. One or more of these bursting parameters are subject to spatially controlled regulation.
In Drosophila development, information encoded in a handful of maternally deposited protein gradients propagates through increasingly complex layers of interacting genes, culminating in the specification of the adult body plan. A priori, there are several distinct regulatory strategies at the single-cell level capable of generating spatially differentiated patterns of cytoplasmic mRNA each with distinct implications for the nature of the underlying molecular processes at play. The average rate of transcription is mainly modulated across the embryo by tuning the frequency of transcriptional bursting. It has remained unclear whether this modulation of the rate of transcription (and thereby mRNA production) is the dominant modality by which input concentrations of transcription factors drive the formation of patterns of gene expression or whether, instead, it is simply the most readily apparent mechanism among multiple distinct control strategies.
While it is widely appreciated that genes are expressed for discrete windows of time over the course of development, in the case of this eve stripe 2 reporter—this binary transcriptionally engaged/quiescent logic is actively regulated by transcription factors to drive pattern formation. Important is the temporal component of transcriptional regulation in specifying developmental outcomes. The limited readout time imposed by short nuclear cycles in early Drosophila development places strict constraints on the kinds of regulatory architecture that could be responsible for driving observed patterns of hunchback gene expression. The pioneer factor Zelda plays a key role in regulating both the timing and probability of transcriptional activation following mitosis. Our work complements these previous observations by exploring yet another aspect of the interplay between timing and transcriptional regulation.
The Drosophila Pioneer Factor Zelda Modulates the Nuclear Microenvironment of a Dorsal Target Enhancer to Potentiate Transcriptional Output April 22, 2019 2
1. https://www.pnas.org/content/117/2/836
2. https://www.sciencedirect.com/science/article/pii/S0960982219303173


