Evolution of Bacteria
Why are there numerous life forms that are far more complex than the minimal, simplest device for replication? We cannot know “for sure” what these minimally complex devices are, but there are excellent candidates —namely, the simplest autotrophic bacteria and archaea, such as Pelagibacter ubique or Prochlorococcus sp. These organisms get by with about 1,300 genes without using any organic molecules, and generally without any dependence on other life forms. Incidentally, these are also the most “successful” organisms on Earth. 8
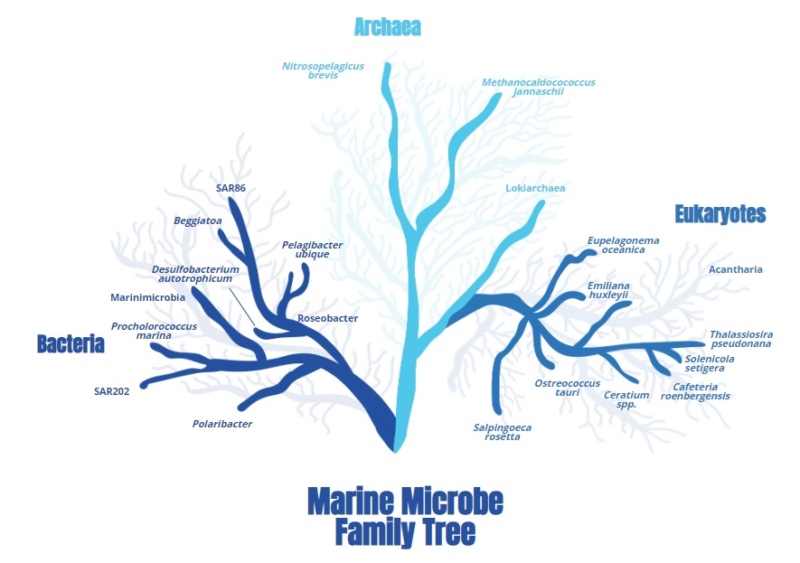
A microbe is a microscopic organism, and it is not necessarily within a unified group that is closely related. In fact, microbes exist in all three domains of life. There are microbes that are bacteria, microbes that are archaea, and microbes that are part of the more complex domain that plants and animals are a part of—the eukaryotes. 9
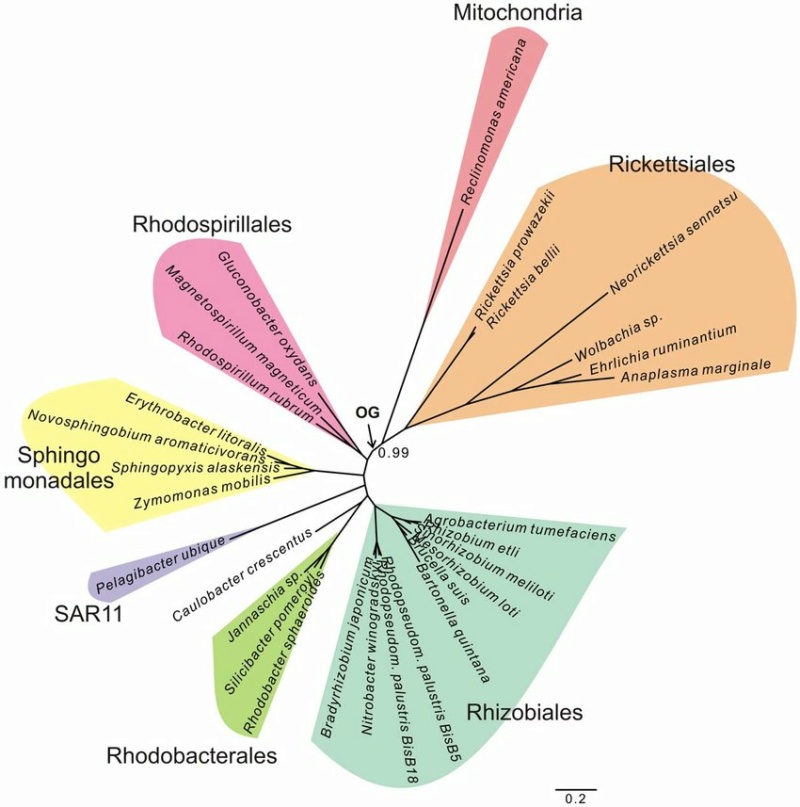
Ca. Pelagibacter ubique is placed with free-living Alphaproteobacteria in a phylogenetic tree inferred from concatenated protein sequences. A phylogenetic inference of the alphaproteobacterial tree topology based on a concatenated alignment of 42 mitochondrially encoded proteins reveals a well-supported alphaproteobacterial phylogeny, in which the SAR11 clade, here represented by Ca. Pelagibacter ubique is placed among the free-living alphaproteobacteria rather than with the Rickettsiales. The tentative placement of the outgroup (OG) is indicated with an arrow. 10
Closest to the universal common ancestor
When the quest arises about the first living organism, and the origin of life, a last universal common ancestor ( LUCA ) is proposed, which is a hypothesized construct, by doing phylogenetic analysis, and attempting to analyze genome data from the three domains of life, and from that, a common ancestor is inferred. Proposals of a Luca are, however, just theoretical attempts to reconstruct the history of life, but there is no empirical evidence, that such a construct could actually live. Often, another organism, Mycoplasma genitalium is mentioned as a reference of the threshold of the living from the non-living. Mycoplasma genitalium is held as the smallest possible living self-replicating cell, and as such, used as a reference of the smallest possible living cell. Mycoplasma is, however, a pathogen, an endosymbiont that only lives and survives within the body or cells of another organism ( humans ). As such, it IMPORTS many nutrients from the host organism. The host provides most of the nutrients such bacteria require, hence the bacteria do not need the genes for producing such compounds themselves. Endosymbionts can only survive inside their host cells as they rely on their host (and, in some cases on co-primary endosymbiotic partners) for metabolic and other functions. As such, it does not require the same complexity of biosynthesis pathways to manufacturing these nutrients as a free-living bacterium.
Amino Acid Transport in Mycoplasma
The fact that the minute Mycoplasma cells lack many biosynthetic pathways and depend on the supply of many nutrients from the growth medium may indicate the presence of numerous transport systems in these organisms
https://jb.asm.org/content/jb/95/5/1685.full.pdf
This indicates, that a hypothetic Last Universal Common Ancestor would have had to be much more complex. Amino Acids, for example, were no readily available on the early earth. For example, in the Miller Urey experiment, eight of the 20 amino acids were never produced. Neither in 1953 nor in the subsequent experiments.
A Last universal common ancestor can also be questioned based on the fact that The DNA replication machinery is not homologous in the 3 domains of life. Bacteria and Archaea differ strikingly in the chemistry of their membrane lipids. Sequences of glycolytic enzymes differ between Archaea and Bacteria/Eukaryotes. There are at least six distinct autotrophic carbon fixation pathways. If common ancestry were true, an ancestral Wood–Ljungdahl pathway should have become life's one and only principle for biomass production. There is a sharp divide in the organizational complexity of the cell between eukaryotes, which have complex intracellular compartmentalization, and even the most sophisticated prokaryotes (archaea and bacteria), which do not, and a typical eukaryotic cell is about 1,000-fold bigger by volume than a typical bacterium or archaeon, and functions under different physical principles: free diffusion has little role in eukaryotic cells but is crucial in prokaryotes.
As such, it makes little sense to hypothesize that life began and started from a Last universal common ancestor. Of course, that fact alone puts in check-mate the materialistic hypothesis, and the narrative falls short that life self-organized somehow from a LUCA, and the diversified through evolution. But in order to have a realistic idea, how random chemistry could have self-organized to become alive, the best is to analyze the complexity of one of the smallest known free-living bacteria.
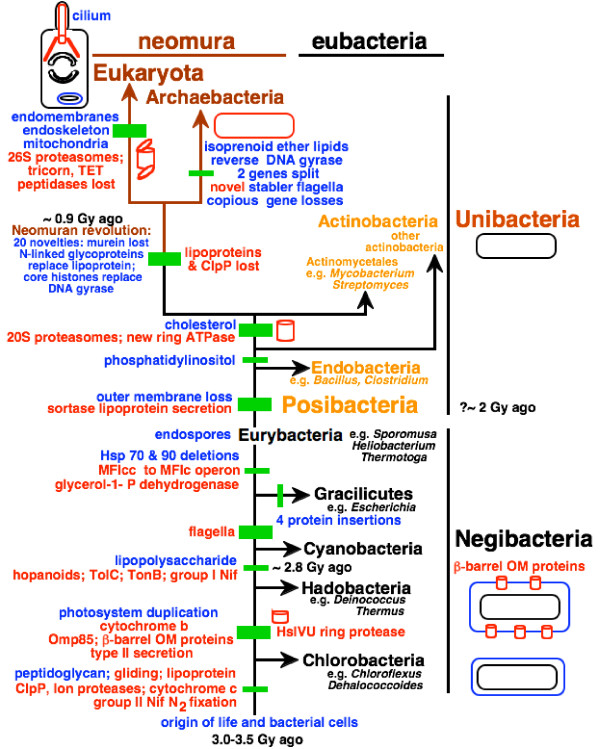
If Chlorobacteria are indeed the earliest diverging bacteria and diverged before oxygenic photosynthesis and two photosystems evolved, they must have been ancestrally anaerobic. Chlorobacteria comprise filamentous 'non-sulphur' green bacteria, e.g. Chloroflexus, Oscillochloris, Chloronema and Heliothrix, which can be photoheterotrophs or photoautotrophs; colourless heterotrophs, including thermophiles (e.g. Thermomicrobium, Herpetosiphon); and chlororespirers (Dehalococcoides). 1 Biological nitrogen fixation is very ancient, dating back to the oldest phylum, Chlorobacteria 3
Chlorobacteria are supposedly the most primitive extant cells 2
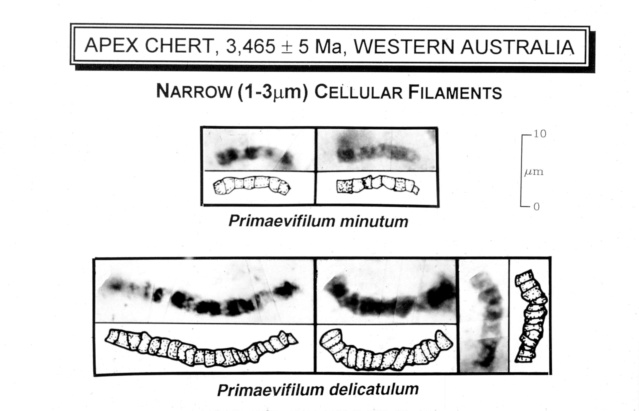
Some of the oldest known fossils from Earth, bacteria fossils from the Apex Chert in Western Australia. Approximately 3.5 billion years ago, this rock was sediment on the floor of an ancient lake or ocean. Bacteria or algae lived on (and in) the sediment and were trapped and preserved in it. These bacteria fossils are only 10–20 micrometers long, so they can only be seen through a microscope. This slide shows 3 of the 11 different kinds of bacteria fossils in the Apex Chert. Most are chains of single cells, forming filaments, very much like bacteria and blue-green algae do today on the floors of modern lakes. 4
An organism in of the phylum Thermotogae was the most recent common ancestor of modern bacteria 5
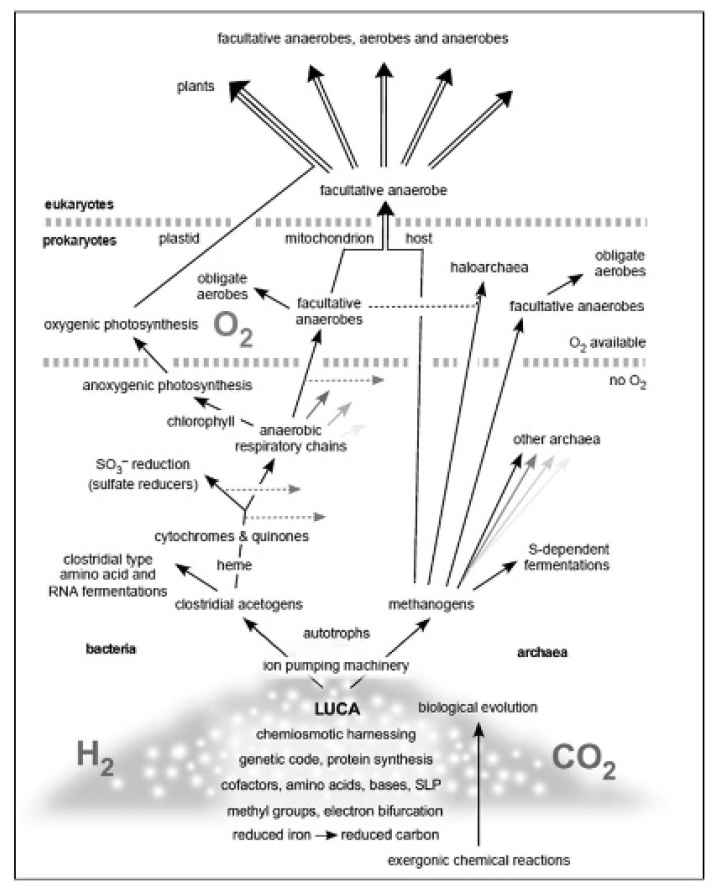
It is hard to piece together what the organisms that belong to environmental sequences are doing in nature based on the information in their genome. One example is the methanogenlike metabolisms that people are finding in marine sediments. Evans et al. (2015) reported new archaeal lineages (Bathyarchaeota) from marine sediment that have the archaeal version of the acetyl-CoA pathway, but have no clear evidence for known forms of archaeal energy-conserving metabolism. He et al. (2016) reported Bathyarchaeota lineages that appear to be performing a very simple and suspected primitive form of energy metabolism that is otherwise only known from bacteria so far—or from archaea under very specific kinds of culture conditions acetogenesis. What kind of carbon and energy metabolism existed at the root of life’s chemical reaction?
Phylogenetic analysis shows that the order Aquificales are at the deep branching position of the Bacteria domain [based on 16S ribosomal RNA (rRNA) sequences 6
It is one of a handful of species in the Aquificae phylum, an unusual group of thermophilic bacteria that are thought to be some of the oldest species in the bacteria domain 7
1. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC1586193/
2. https://royalsocietypublishing.org/doi/10.1098/rstb.2009.0161
3. https://royalsocietypublishing.org/doi/10.1098/rstb.2006.1842
4. https://www.lpi.usra.edu/publications/slidesets/marslife/slide_31.html
5. https://en.wikipedia.org/wiki/Evolution_of_bacteria
6. https://sci-hub.tw/https://link.springer.com/article/10.1007/s00253-007-0893-0
7. https://en.wikipedia.org/wiki/Aquifex_pyrophilus
8. https://reasonandscience.catsboard.com/t2234-the-origin-of-replication-and-translation-and-the-rna-world
9. https://ocean.si.edu/ocean-life/microbes/marine-microbe-tree-life
10. https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0024457
Why are there numerous life forms that are far more complex than the minimal, simplest device for replication? We cannot know “for sure” what these minimally complex devices are, but there are excellent candidates —namely, the simplest autotrophic bacteria and archaea, such as Pelagibacter ubique or Prochlorococcus sp. These organisms get by with about 1,300 genes without using any organic molecules, and generally without any dependence on other life forms. Incidentally, these are also the most “successful” organisms on Earth. 8

A microbe is a microscopic organism, and it is not necessarily within a unified group that is closely related. In fact, microbes exist in all three domains of life. There are microbes that are bacteria, microbes that are archaea, and microbes that are part of the more complex domain that plants and animals are a part of—the eukaryotes. 9

Ca. Pelagibacter ubique is placed with free-living Alphaproteobacteria in a phylogenetic tree inferred from concatenated protein sequences. A phylogenetic inference of the alphaproteobacterial tree topology based on a concatenated alignment of 42 mitochondrially encoded proteins reveals a well-supported alphaproteobacterial phylogeny, in which the SAR11 clade, here represented by Ca. Pelagibacter ubique is placed among the free-living alphaproteobacteria rather than with the Rickettsiales. The tentative placement of the outgroup (OG) is indicated with an arrow. 10
Closest to the universal common ancestor
When the quest arises about the first living organism, and the origin of life, a last universal common ancestor ( LUCA ) is proposed, which is a hypothesized construct, by doing phylogenetic analysis, and attempting to analyze genome data from the three domains of life, and from that, a common ancestor is inferred. Proposals of a Luca are, however, just theoretical attempts to reconstruct the history of life, but there is no empirical evidence, that such a construct could actually live. Often, another organism, Mycoplasma genitalium is mentioned as a reference of the threshold of the living from the non-living. Mycoplasma genitalium is held as the smallest possible living self-replicating cell, and as such, used as a reference of the smallest possible living cell. Mycoplasma is, however, a pathogen, an endosymbiont that only lives and survives within the body or cells of another organism ( humans ). As such, it IMPORTS many nutrients from the host organism. The host provides most of the nutrients such bacteria require, hence the bacteria do not need the genes for producing such compounds themselves. Endosymbionts can only survive inside their host cells as they rely on their host (and, in some cases on co-primary endosymbiotic partners) for metabolic and other functions. As such, it does not require the same complexity of biosynthesis pathways to manufacturing these nutrients as a free-living bacterium.
Amino Acid Transport in Mycoplasma
The fact that the minute Mycoplasma cells lack many biosynthetic pathways and depend on the supply of many nutrients from the growth medium may indicate the presence of numerous transport systems in these organisms
https://jb.asm.org/content/jb/95/5/1685.full.pdf
This indicates, that a hypothetic Last Universal Common Ancestor would have had to be much more complex. Amino Acids, for example, were no readily available on the early earth. For example, in the Miller Urey experiment, eight of the 20 amino acids were never produced. Neither in 1953 nor in the subsequent experiments.
A Last universal common ancestor can also be questioned based on the fact that The DNA replication machinery is not homologous in the 3 domains of life. Bacteria and Archaea differ strikingly in the chemistry of their membrane lipids. Sequences of glycolytic enzymes differ between Archaea and Bacteria/Eukaryotes. There are at least six distinct autotrophic carbon fixation pathways. If common ancestry were true, an ancestral Wood–Ljungdahl pathway should have become life's one and only principle for biomass production. There is a sharp divide in the organizational complexity of the cell between eukaryotes, which have complex intracellular compartmentalization, and even the most sophisticated prokaryotes (archaea and bacteria), which do not, and a typical eukaryotic cell is about 1,000-fold bigger by volume than a typical bacterium or archaeon, and functions under different physical principles: free diffusion has little role in eukaryotic cells but is crucial in prokaryotes.
As such, it makes little sense to hypothesize that life began and started from a Last universal common ancestor. Of course, that fact alone puts in check-mate the materialistic hypothesis, and the narrative falls short that life self-organized somehow from a LUCA, and the diversified through evolution. But in order to have a realistic idea, how random chemistry could have self-organized to become alive, the best is to analyze the complexity of one of the smallest known free-living bacteria.

If Chlorobacteria are indeed the earliest diverging bacteria and diverged before oxygenic photosynthesis and two photosystems evolved, they must have been ancestrally anaerobic. Chlorobacteria comprise filamentous 'non-sulphur' green bacteria, e.g. Chloroflexus, Oscillochloris, Chloronema and Heliothrix, which can be photoheterotrophs or photoautotrophs; colourless heterotrophs, including thermophiles (e.g. Thermomicrobium, Herpetosiphon); and chlororespirers (Dehalococcoides). 1 Biological nitrogen fixation is very ancient, dating back to the oldest phylum, Chlorobacteria 3
Chlorobacteria are supposedly the most primitive extant cells 2

Some of the oldest known fossils from Earth, bacteria fossils from the Apex Chert in Western Australia. Approximately 3.5 billion years ago, this rock was sediment on the floor of an ancient lake or ocean. Bacteria or algae lived on (and in) the sediment and were trapped and preserved in it. These bacteria fossils are only 10–20 micrometers long, so they can only be seen through a microscope. This slide shows 3 of the 11 different kinds of bacteria fossils in the Apex Chert. Most are chains of single cells, forming filaments, very much like bacteria and blue-green algae do today on the floors of modern lakes. 4
An organism in of the phylum Thermotogae was the most recent common ancestor of modern bacteria 5

It is hard to piece together what the organisms that belong to environmental sequences are doing in nature based on the information in their genome. One example is the methanogenlike metabolisms that people are finding in marine sediments. Evans et al. (2015) reported new archaeal lineages (Bathyarchaeota) from marine sediment that have the archaeal version of the acetyl-CoA pathway, but have no clear evidence for known forms of archaeal energy-conserving metabolism. He et al. (2016) reported Bathyarchaeota lineages that appear to be performing a very simple and suspected primitive form of energy metabolism that is otherwise only known from bacteria so far—or from archaea under very specific kinds of culture conditions acetogenesis. What kind of carbon and energy metabolism existed at the root of life’s chemical reaction?
Phylogenetic analysis shows that the order Aquificales are at the deep branching position of the Bacteria domain [based on 16S ribosomal RNA (rRNA) sequences 6
It is one of a handful of species in the Aquificae phylum, an unusual group of thermophilic bacteria that are thought to be some of the oldest species in the bacteria domain 7
1. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC1586193/
2. https://royalsocietypublishing.org/doi/10.1098/rstb.2009.0161
3. https://royalsocietypublishing.org/doi/10.1098/rstb.2006.1842
4. https://www.lpi.usra.edu/publications/slidesets/marslife/slide_31.html
5. https://en.wikipedia.org/wiki/Evolution_of_bacteria
6. https://sci-hub.tw/https://link.springer.com/article/10.1007/s00253-007-0893-0
7. https://en.wikipedia.org/wiki/Aquifex_pyrophilus
8. https://reasonandscience.catsboard.com/t2234-the-origin-of-replication-and-translation-and-the-rna-world
9. https://ocean.si.edu/ocean-life/microbes/marine-microbe-tree-life
10. https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0024457


