Key components involved in Hedgehog signalling
-The Hedgehog ligand, Hedgehog (Hh)
Initiates signal transduction of the Hedgehog pathway
-The Hedgehog ligand receptor, Patched (PTCH)
Normally suppresses the activity of SMO
-I-hog receptors
-The cell surface signal transducer, Smoothened (SMO)
Normally suppressed by PTCH, preventing its activation of the Hedgehog signalling cascade
-A microtubule-associated complex of Cos2/Kif7, Fused (Fu)
-Suppressor of Fused (Sufu)
-PKA
-GSK3
-CK1
-Key effector Ci/Gli zink finger transcription factor
-The downstream effectors,the Gli transcription factors
[ltr]Cytosolic complex of proteins including Suppressor of Fused (SuFu) and the Gli family of transcription factors. Activation leads to expression of specific genes that promote cell proliferation and differentiation[/ltr]
-the transcription factor Cubitus interruptus (Ci).
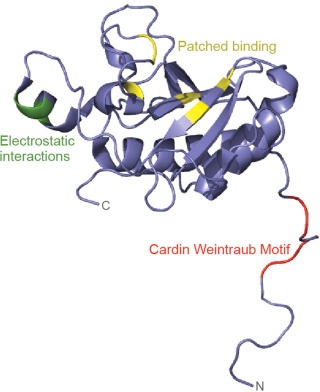
All of these components are also present in the cnidarians. The ligands of the Hedgehog pathway are unique proteins which can auto-catalyse cleavage of the N-terminal signalling fragment from the C-terminal Intein domain. The N-terminal undergoes lipid modification, including covalent attachment of cholesterol; this modification is critical for Hedgehog’s ability to signal across long distances. A twelve-transmembrane domain protein, Patched, which belongs to a family of sterol-sensing proteins, is the core receptor of the cholesterol-linked Hedgehog molecule, and a single-transmembrane protein I-hog acts as a co-receptor.
Patched (Ptc) is a conserved 12-pass transmembrane protein receptor that plays an obligate negative regulatory role in the Hedgehog signaling pathway in insects and vertebrates. Patched is an essential gene in embryogenesis that is important for proper segmentation in the fly embryo, mutations in which may be embryonic lethal.
Formation of Hh-Patched-Ihog complex results in phosphorylation of a cytoplasmic tail of the key mediator of the pathway: a seven-transmembrane protein Smoothened (which is related to the Wnt receptors, Frizzled).
Mechanisms and functions of Hedgehog signalling across the metazoa 1
Structure and properties of Hedgehog proteins
Hedgehog proteins are synthesized as approximately 45 kDa pro-proteins that undergo autocleavage to yield two similarly sized fragments8 : an amino-terminal polypeptide (HH-N), which is characterized by a signal sequence and a highly conserved ‘Hedge’ domain, and a carboxy-terminal polypeptide (HH-C), which contains the highly conserved ‘Hog’ domain9 (FIG. 1a).
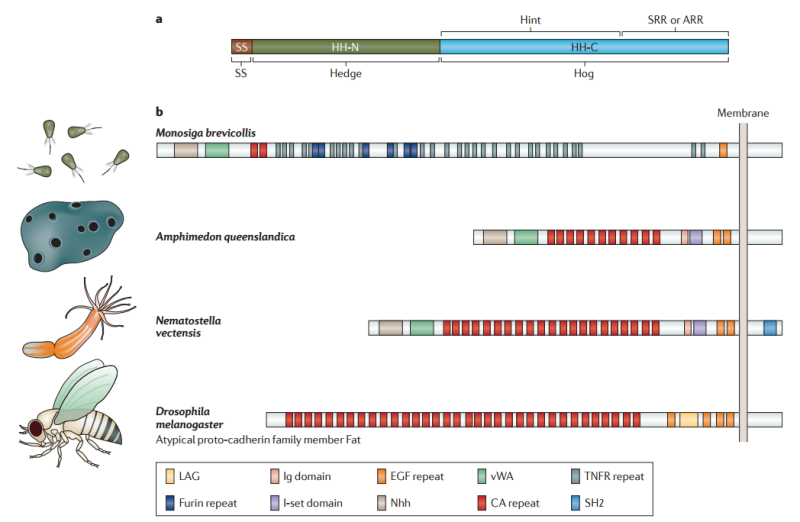
Figure 1 | Structure of Hedgehog and Hedgehog-related proteins.
a | The full-length Hedgehog (HH) protein is comprised of two distinct domains: the amino-terminal ‘Hedge’ domain (shown in dark green) and the carboxy-terminal ‘Hog’ domain (shown in blue), both of which are also found in proteins other than members of the HH family. The Hedge domain is preceded by a signal peptide sequence (SS; shown in brown). The Hog domain itself can be separated into two regions; the first two-thirds share similarity to self-splicing inteins, and this module has been named ‘Hint’, whereas the C-terminal one-third binds cholesterol in HH proteins and has been named the sterol-recognition region (SRR). In other Hog domain-containing proteins this region is referred to as the adduct recognition region (ARR), as the nature of the adduct is not known. The Hog domain promotes autocleavage to release the N-terminal Hedge domain (HH-N).
b | Hedge domain-containing proteins predate HH and are found in protozoa and metazoa. The choanoflagellate Monosiga brevicollis has a large Hedge domain-containing transmembrane protein that also contains a von Willebrand A domain (vWA), two cadherin repeats (CA repeats), multiple tumour necrosis factor receptor repeats (TNFR repeats), as well as single immunoglobulin (Ig), immunoglobulin I-set (I-set) and epidermal growth factor (EGF)-like repeats14; the Hedge domain is designated Nhh. Proteins with a similar domain organization, though with an expansion of the CA repeats at the expense of the TNFR repeats, are encoded by the so-called hedgling gene in the basal metazoan, Amphimedon queenslandica (sponge)15 and the Cnidarian, Nematostella vectensis15. Interestingly, the hedgling structure resembles that of the atypical proto-cadherin family members, including the Fat and Dachsous proteins, which have important roles in planar cell polarity in Drosophila melanogaster154. LAG, Laminin A-G motif; SH2, SH2 motif.
All of the signalling activity of HH proteins is vested in the secreted HH-N fragment, which has the unusual property of being covalently coupled to cholesterol at its C-terminal end 3 . The only known function of the Hog domain is to promote the autocleavage reaction, a process that resembles the protein-splicing activity of inteins and requires the ‘Hint’ region, which forms part of the Hog domain 10. The cleavage reaction is driven by a nucleophilic attack on a highly conserved GCF motif in which cholesterol, recruited by the sterol recognition region (SRR) of the Hog domain, serves as the electron donor 3 . The Hedge and Hog domains have ancient origins. Although Hedgehog proteins are the only proteins known to be conjugated to cholesterol in this way, the role of the Hog domain in processing secreted proteins seems to be an ancient one: Hog domain proteins have been identified in a wide variety of organisms, including red algae, dinoflagellates and mosses, as well as throughout the metazoa (although not in higher plants) 11. In the choanoflagellate Monosiga ovata, the protist that is most closely related to the metazoa, the ‘Hoglet’ gene encodes a protein with a C-terminal Hog domain that is coupled to an N-terminal domain with sequence similarity to cellulose-binding proteins 12. However, in other organisms Hog domains have been found to be associated with an assortment of unrelated N-terminal domains. Only in the eumetazoa does the association of the Hedge and Hog domains in the form of Hedgehog proteins first appear 11
The Hedge domain, which has a striking structural similarity to the Streptomyces albus metalloproteinase d,d-carboxypeptidase 13, is also found in Monosiga spp., in the N-terminal extracellular domain of a large transmembrane protein 14 (FIG. 1b). A protein with a similar domain organization is encoded by the so-called Hedgling gene in the basal metazoan Amphimedon queenslandica (Porifera)15. The Cnidarian, Nematostella vectensis (the sea anenome) also has a Hedgling gene, as well as two true Hedgehog genes16; by contrast, no Hedgling genes have been reported in any bilaterian species. Thus, it seems likely that Hedgehog proteins first arose in the common ancestor of the Cnidarians and the bilateria more than 650 million years ago through the combination of domains that were present in preexisting proteins, and that the Hedgling gene was subsequently lost from the bilateria 11. Most bilaterians have been shown to possess at least one Hedgehog gene, with the genome expansions in vertebrates giving rise to three genes in amniotes (Desert Hedgehog (Dhh), Indian Hedgehog (Ihh) and Sonic Hedgehog (Shh)) and four or five (Dhh, Ihha, Ihhb, Shha and Shhb) in different teleost species 17. A notable exception is C. elegans, which possesses multiple Hog domain-containing proteins but lacks a true Hedgehog gene2 . By contrast, some other nematode species have retained a Hedgehog gene, though, like C. elegans and its closer relatives, they have lost some of the components of the core signal transduction pathway 11 (described below).
Lipid modifications control the release and movement of HH.
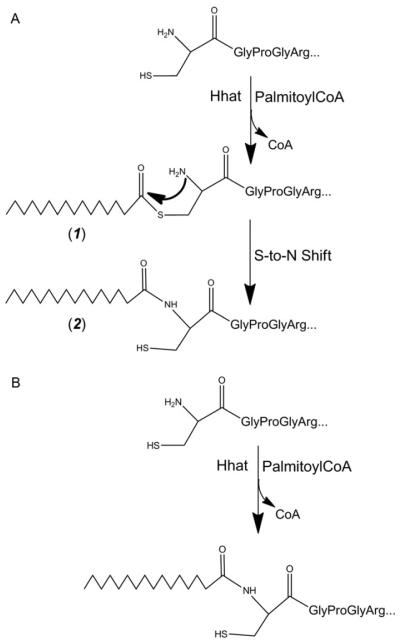
In addition to the covalent coupling of cholesterol to their C-termini, HH-N proteins also undergo palmitoylation at their N-termini, a modification that is promoted by an acyl transferase encoded in D. melanogaster by the skinny hedgehog (ski) gene18 and in vertebrates by its orthologue, HHAT19 (FIG. 2).
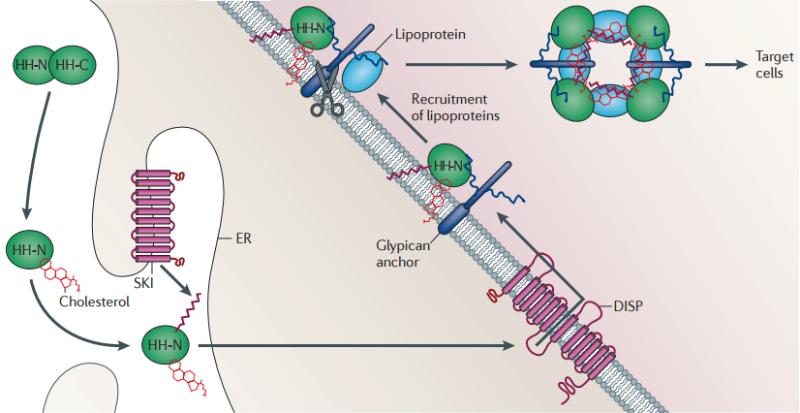
Figure 2 | Lipid modification and release of the Hedgehog ligand.
Following its translation, full-length Hedgehog (HH) undergoes autoproteolysis in the endoplasmic reticulum (ER)155, resulting in its covalent coupling to cholesterol. The amino-terminal ‘Hedge’ domain (HH-N) fragment is further modified through N-terminal palmitoylation that is promoted by the transmembrane acyl transferase Skinny hedgehog (SKI). Release of this doubly lipid-conjugated form of HH requires the activity of the multipass transmembrane protein Dispatched (DISP), which probably transports the protein across the plasma membrane. Once on the outer surface of the cell, HH remains associated with the lipid bilayer and interacts with the heparan sulphate moieties of the glypicans that are encoded by the dally and dally-like genes (in Drosophila melanogaster). These are also thought to recruit the apolipoprotein lipophorin, which, together with HH, becomes assembled into lipoprotein particles. Release of these particles might be mediated by the phospholipase C-like Notum, which cleaves the GPI anchors from the glypicans (indicated by scissors).
This dual lipid modification of the HH signalling protein has important effects on its properties, both enhancing its membrane association 20 and potentiating its secretion and range of action. The secretion of lipidated HH-N requires the activity of a large multipass transmembrane protein, Dispatched (DISP) 21. In mutant animals lacking DISP function, HH accumulates in producing cells and all HH-elicited responses are lost, except in cells that are immediately adjacent to signal producers 21–24. Truncated proteins that are engineered to mimic HH-N but which lack the cholesterol moiety can override this requirement for DISP; however, the range and potency of such cholesterol-free HH-N is severely compromised compared with the normal protein, suggesting that the modification is crucial for the extracellular movement of the signal following secretion 25,26. Consistent with this, the biochemical analysis of vertebrate SHH expressed in tissue culture cells indicates that lipid modification promotes the formation of freely diffusible multimeric complexes 27.
In D. melanogaster, studies of HH signalling in imaginal discs have shown that the cholesterol adduct is essential for the incorporation of HH-N into lipoprotein particles that seem to mediate its long-range transport across epithelia28 (FIG. 2). The assembly of these HH signalling entities is promoted by interaction with lipophorin, an apo-lipoprotein that circulates in the haemolymph of the fly 29. Lipophorin is recruited to HH-N-secreting cells by its interaction with the heparin sulphate moieties of the glypicans Dally and Dally-like29. These proteoglycans, which can also interact with HH30,31, localize to the apical surfaces of epithelial cells via GPI anchors, the cleavage of which seems to be required for effective long-range HH signalling32. Although the properties of the Dally and Dally-like mutants can been subject to differing interpretations29,32,33, one appealing possibility is that glypicans promote the assembly of HH-N–lipophorin particles at the plasma membrane and that cleavage of their GPI anchor facilitates the release and dispersal of HH from producing cells (FIG. 2). Notably, HH-N also localizes to the basolateral membrane of secreting and receiving cells in imaginal discs, leading to the proposal by some authors that this represents the major route of signal secretion and spread34,35. Experimental generation of HH-N ‘traps’ in groups of responding cells, however, suggests that the range of the basally secreted protein is fairly limited32: this raises the interesting possibility that basal HH-N principally acts as a short-range signal, whereas apically released HH-N acts as a long-range signal. In line with this, apically localized HH-N can be observed across the imaginal disc epithelium in a graded distribution that mirrors the functionally defined gradient of signalling activity32.
Heparin binding induces Ihog dimerization and is required to mediate high-affinity interactions between Ihog and Hh. 3 We also present crystal structures of a Hh-binding fragment of Ihog, both alone and complexed with Hh. Heparin is not well ordered in these structures, but a basic cleft in the first FNIII domain of Ihog (IhogFn1) is shown by mutagenesis to mediate heparin binding. These results establish that Hh directly binds Ihog and provide the first demonstration of a specific role for heparin in Hh responsiveness. Hh is secreted but undergoes two lipid modifications that restrict its free diffusion and facilitate transport to appropriate target sites.
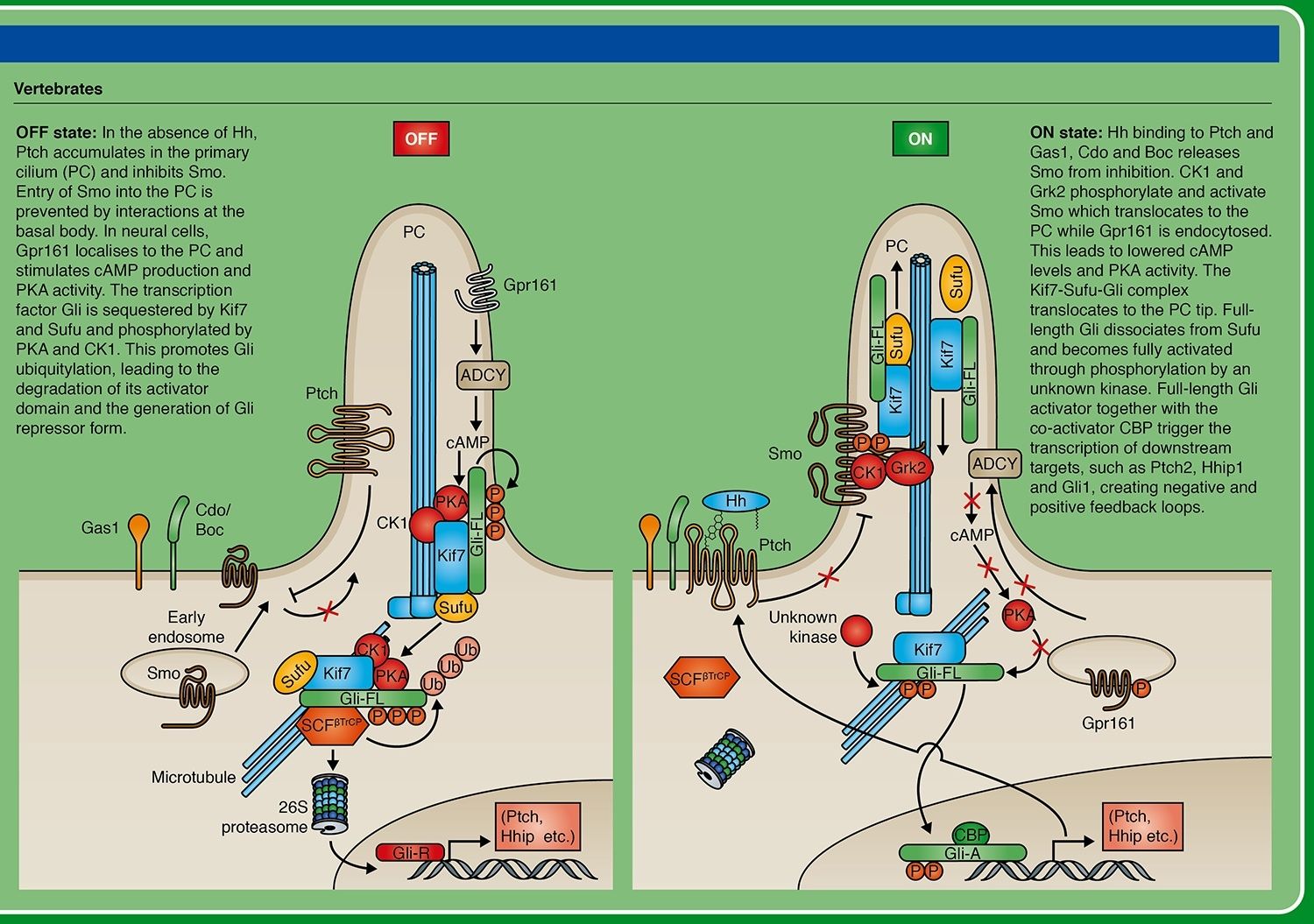
Phosphorylated Smoothened translocates from micro-vesicles, where it remained in the absence of Hh signal, to the plasma membrane (in vertebrates, to the plasma membrane of the primary cilium). From there, it promotes dissociation of the microtubule-associated complex of several proteins (including kinases PKA, GSK3 and CK1) which in the absence of signal phosphorylated the transcription factor Ci/Gli. Phosphorylated Ci/Gli undergoes proteolytic cleavage, which removes its C-terminal activator domain and generates a transcriptional repressor form (Ci/GliR). Presence of the Hedgehog signal, through dissociation of the kinases complex, results in accumulation of non-phosphorylated Ci/Gli and its subsequent (but not fully understood) processing resulting in formation of an activator form (Ci/GliA) and allowing strong activation of the target genes.
The hedgehogs were first identified because mutations of the gene in Drosophila disrupted the segmentation pattern and made the larvae look like hedgehogs. Sonic hedgehog is very important for the dorsoventral patterning of the neural tube and for anteroposterior patterning of the limbs. Indian hedgehog is important in skeletal development. The full-length hedgehog polypeptide is an autoprotease, cleaving itself into an active N-terminal and an inactive C-terminal part. The Nterminal fragment is normally modified by covalent addition of a fatty acyl chain and of cholesterol, which are needed for full activity. The hedgehog receptor is called patched, again named after the phenotype of the gene mutation in Drosophila. This is of the G-protein-linked class. It is constitutively active and is repressed by ligand binding. When active it represses the activity of another cell membrane protein, smoothened, which in turn represses the proteolytic cleavage of Gli-type transcription factors. Full-length Gli factors are transcriptional activators that can move to the nucleus and turn on target genes, but the constitutive removal of the C-terminal region makes them into repressors. In the absence of hedgehog, patched is active, smoothened inactive, and Gli inactive. In the presence of hedgehog, patched is inhibited, smoothened is active, and Gli is active (Fig. A.4d). Activation of protein kinase A also represses Gli and
hence antagonizes hedgehog signaling.
The Hedgehog (Hh) signaling pathway has numerous roles in the control of cell proliferation, tissue patterning, stem cell maintenance and development. The primary cilium is an important center for transduction of the Hedgehog signal in vertebrates. In Hh's absence, the Ptch receptor localizes to the cilium, where it inhibits Smo activation. Gli proteins are phosphorylated by PKA, CKI and GSK3B and partially degraded into truncated Gli repressor form (GliR) that suppresses Hh target gene transcription in the nucleus. In Hh's presence, Ptch disappears from the cilium, and activated Smo contributes to the translocation of the protein complex Gli, Sufu, Kif7 to ciliary tip, where Gli dissociates from the negative regulator Sufu. The production of Gli activator form (GliA) occurs and the increased nuclear accumulation of GliA results in activation transcription of Hh target genes.
Originally defined through genetic analysis in D. melanogaster 1 , the components of the HH signalling pathway have subsequently been functionally characterized in a number of vertebrate species — principally, mouse, zebrafish and human — and have also been identified through genome sequence analyses in species from a wide range of phyla (FIG. 3). These studies have revealed a high level of conservation of the ‘core’ components of the signal transduction pathway that is likely to extend across the eumetazoa, as well as some variation in their deployment that has arisen over the course of evolution.
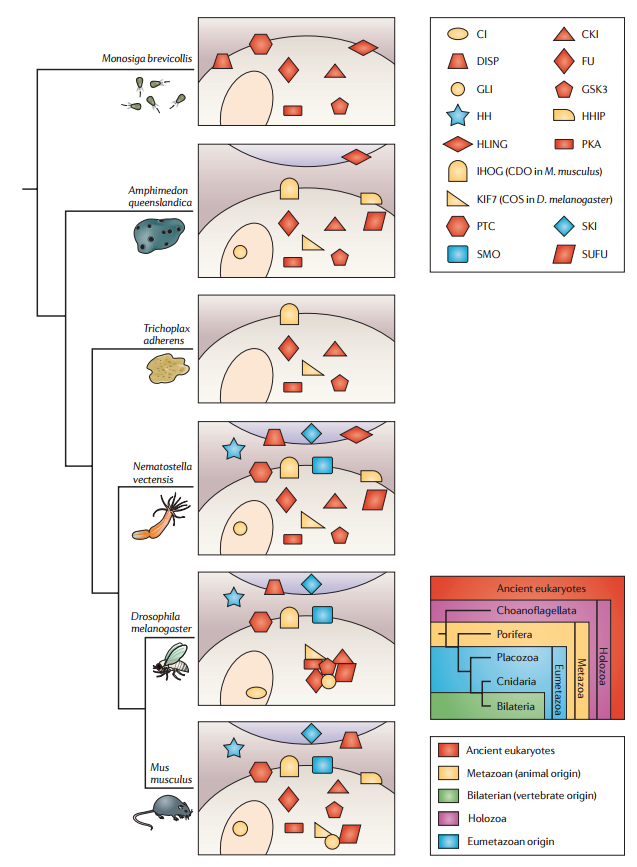
Figure 3 | Conservation of individual molecules across species with phylogenetic representation.
Genomic inventory of Hedgehog (HH) pathway components in species representative of different phyla. The components are colour-coded to indicate their likely origin and are distributed between receiving cells (shaded beige, nuclei shown as ovals) and signalling cells (shaded purple, top of panels). Interactions established on the basis of genetic and/or biochemical data are indicated by the clustering of components. CKI, casein kinase 1; COS, Costal; DISP, Dispatched; FU, Fused; GSK3, glycogen synthase kinase 3; HHIP, Hedgehog-interacting protein; HLING, hedgling; IHOG, Interference hedgehog; PKA, protein kinase A; PTC, Patched; SKI, Skinny hedgehog; SMO, Smoothened; SUFU, Suppressor of fused
Hedgehog signaling in animal development: paradigms and principles 2
The last decade has seen extraordinary progress in deciphering the roles and mechanisms of action of Hedgehog proteins. From models in the early 1990s of a short-range signal regulating pattern in the ectoderm of the Drosophila embryo, Hh proteins are now recognized as acting both locally and at long range to regulate a plethora of processes in vertebrate as well as invertebrate development. At every step in the unraveling of the signaling pathway there have been surprises, a fact that bears ample testimony to the power of genetic analysis in uncovering novel paradigms and principles. The first of these is the unusual autoprocessing that generates the active Hh ligands, a process that simultaneously couples them to cholesterol. As we have discussed above, this unique lipid modification contributes to some key properties of the Hh signal, mediating its controlled release and movement from its source. The process of cell-to cell transport depends on two other components that appear to be dedicated to Hh signaling: Dispatch-mediated release of Hh-Np from the sending cell and Toutvelu-dependent trafficking across the target field. Cholesterol coupling may also ensure that Hh ligands are concentrated in membranes, increasing the likelihood of ligand/receptor interaction. Furthermore, the cholesterol anchor may target Hh ligands to membrane subdomains that also localize Ptc or Hip1, thereby facilitating either signaling or its termination by ligand sequestration. Robust negative-feedback mechanisms are a hallmark of most signaling pathways, and it is clear that in both the fly and the mouse, effective sequestration of Hh by Ptc is dependent on its cholesterol linkage. A second lipid modification, palmitoylation of the N terminus, also plays a key role, but most likely not in membrane retention or movement. Rather, increasing the overall hydrophobicity of this part of the protein some how enhances its ability to inactivate Ptc. Determining the structural basis of this effect will be of key importance in understanding the dose-dependent effects of Hh ligands. A further distinguishing feature of the Hedgehog pathway is its mechanism of receptor-mediated activation. In most cases, extracellular signals elicit their effects by binding to and activating a membrane-anchored receptor that, in turn, activates intracellular components of the pathway. Hh proteins, in contrast, act by repressing their receptor, Ptc, which, in turn, controls the expression of Hh target genes by repressing the activity of Smo. What is the logic of this unusual mechanism? Presently the answer is unclear, but most likely it relates to the peculiarities of Smo activation through some Ptc-dependent intracellular trafficking process, a feature that Ptc might share with SCAP, another SSD-containing regulatory factor. The recent discovery of the RABprotein encoded by the opb gene apparently dedicated to this process, strengthens this view. Future analysis of the subcellular behavior of Ptc and Smo should yield some important new insights into this enigmatic process.
Finally, we have described the unusual way in which Hh signaling elicits its effects at the transcriptional level by altering the sign of a bifunctional transcriptional regulator. In Drosophila, the absence of Hh ligand allows the cleavage of the Ci protein, converting it to a repressor form that can bind target genes to block their transcription. Derepression of Smo, in contrast, inhibits this cleavage and promotes the nuclear import of activated full-length Ci, leading to the transcription of Hh target genes. This highly economic process is further exemplified by the finding that most, if not all, Hh signaling is mediated through Ci in Drosophila. Why, then, do vertebrates use three distinct Gli proteins as transcriptional effectors? One simple explanation could be that each operates similarly, but that gene duplication and the acquisition of new regulatory motifs have led to new tissues that incorporate Hedgehog signaling. Yet neither the expression patterns nor the activities of the different Gli proteins suggest this to be the case. Rather, there appears to have been a partial separation of repressor and activator activities into individual Gli proteins. At least one advantage of this elaboration would be to allow more complex responses within a target field; thus, the response of cells to Hh signaling would be dependent not only on the levels of ligand to which they are exposed but also on the particular repertoire of Gli genes that they express. Further analysis of the in vivo regulation of Gli proteins and of their binding specificities for different Hedgehog targets should help illuminate this aspect of the pathway. It is striking that so much of what is known about this fascinating signaling mechanism to date has been gleaned from genetic analysis, be it in flies or mice. Yet whereas genetics has provided an elaborate framework for our understanding, future progress will require a concerted effort to dissect the signaling process at the biochemical and cell biological levels. The great advances that have been made in identifying Hh-dependent processes and describing the consequences of their activities must now be matched by elucidating the ways in which Hh activities elicit these different cellular responses. Characterization of the multimeric complex that regulates Ci activity has provided a solid basis for this analysis, but many questions remain, not least how Ptc and Smo interact and how Hh binding modulates their interaction. The coming years promise to be at least as revealing as the last.
Hedgehog 4
Hedgehog function
The hedgehog (hh) mutant and gene were originally characterized in Drosophila. Hh proteins are widely conserved within the animal kingdom. Hh signaling is involved in diverse developmental processes, and when misregulated, has been implicated in disease. Loss of Hh signaling in wing discs impedes growth and patterning in both A and P compartments and gives rise to severely reduced wings. Hh acts directly to pattern the central region of the wing. The long-range effects of reduced Hh are secondary consequences due to loss of Dpp expression along the AP compartment boundary in the wing and loss of Dpp and Wg in the leg. Ectopic expression of Hh in A cells at a distance from the AP boundary or activation of signaling by removing Hh pathway inhibitors such as patched (Ptc), protein kinase A (PKA), Costal- 2 (Cos-2), or Slimb (Slmb) causes dramatic reorganization of anterior pattern. Ectopic expression of Hh can lead to complete mirror image duplication of anterior structures. This shows the importance of localized Hh signaling during wing development. Asymmetry is critical to the function of the H patterning system. P cells, which express Hh, are not capable of transducing the Hh signal. They do not express the transcription factor Ci (due to repression by En). Anterior cells that do not receive Hh, process the full-length activator Ci protein (Ci155 or CiA) into a transcriptional repressor form Ci75 or CiR. Cells close to the boundary that receive Hh input stabilize Ci155, leading to the induction of Hh target genes. The distance from the Hh source determines which Hh target genes are activated. Hh can therefore be defined as a morphogen. Hh regulates dpp, wg, ptc, en, and collier (synonym: knot) expression in a concentrationdependent manner.
Hedgehog production
Hh protein is produced as a precursor that undergoes autoproteolytic intramolecular cleavage by an intein-like mechanism. This generates a 20 kDa N terminal (Hh-N) and a 25 kDa C terminal fragment (Hh-C). The enzymatic activity required for the cleavage reaction is contained within the C terminal domain, while the N terminal domain accounts for all known signaling activity . During the cleavage
reaction, a cholesterol moiety is covalently attached to the C terminal part of Hh-N. Cholesterol modification may be responsible for the observed tight cell association of Hh-N to Hh producing
cells. The range over which Hh-N can move and signal is greatly extended when the cholesterol modification is prevented. In addition, Hh is palmitoylated at its N terminus. This lipid modification is also essential for Hh function. In the absence of the acyltransferase Skinny Hedgehog (Ski), Hh lacks the N terminal palmitate residue and cannot fulfill its embryonic and larval patterning activities.
Hedgehog movement
How do these posttranslational modifications impact on Hh movement? The cholesterol and palmitate moieties presumably bind Hh to the cell surface and might be expected to preclude diffusion. Yet Hh protein and Hh activity can be detected many cell diameters away from the cells where it is produced. The release of cholesterol-modified Hh-N from the producing cells depends on the activity of the protein Dispatched (Disp) in Hh-producing cells
. The disp mutant cells appear to produce, process, and modify Hh normally but are unable to release Hh-N and instead accumulate the protein to high levels. Intriguingly, disp is predicted to encode a sterol-sensing domain
(SSD)-containing 12-pass transmembrane protein with sequence homology to the Hh-binding receptor protein Ptc. Ptc has been shown to limit Hh movement,in part, by targeting Hh for internalization and degradation. Thus, both the sending and the receiving cells require the activity of an SSD-containing protein to regulate the movement of Hh-N. Receiving cells need Ptc to sequester cholesterol modified Hh-N while sending cells need Disp to release it. How exactly Disp works in Hh release remains to be determined. It is also not known how Hh moves across tissues. This may be a case where exovesicle (argosome)-mediated movement may prove to be
important. Once Hh is released, its ability to move is influenced by heparan sulfate proteoglycans (HSPGs). The toutvelu (ttv) gene encodes an enzyme needed for HSPG biosynthesis and its activity is needed to allow movement of cholesterol-modified Hh-N in A cells. HSPGs are highly O-glycosylated proteins found abundantly at the cell surface and the extracellular matrix. HSPGs interact with a variety of extracellular proteins such as growth factors, proteases, protease inhibitors, and adhesion molecules. Through these interactions, they participate in many events during cell adhesion, migration, proliferation, and differentiation. Biochemical analyses have indicated that Hh is a heparin-binding protein . How exactly Ttv and HSPGs affect the distribution of Hh-N is still unknown. Hh-N may require HSPGs for its stability in the extracellular space. Alternatively, binding of Hh-N to HSPGs could promote its release from producing cells, prevent its reinsertion into the membrane or its sequestration by Ptc. One could speculate that transport of lipid-modified Hh depends on a yet unidentified carrier protein able to mask the lipid moiety. The possibility of a connection between such a protein and the HSPGs remains to be explored.
Direct patterning activity of Hedgehog
Although the largest morphological changes observed in wings lacking or ectopically activating Hh signaling are caused by changes in the expression of the Hh target gene Dpp, Hh also has a more direct effect on patterning. Hh acts directly to pattern the central region of the wing, more specifically the region between the veins 3 and 4. This short-range patterning activity of Hh is at least in part mediated through the local induction of En and the transcription factor Collier (Col). Another important function of Hh, mediated through a Ci-dependent but thus far unidentified target gene, is preventing A and P cells from mixing, thus maintaining the AP compartment boundary.
Hh is palmitoylated at its N terminus. This lipid modification is also essential for Hh function. In the absence of the acyltransferase Skinny Hedgehog (Ski), Hh lacks the N terminal palmitate residue and cannot fulfill its embryonic and larval patterning activities.
both the sending and the receiving cells require the activity of an SSD-containing protein to regulate the movement of Hh-N
The toutvelu (ttv) gene encodes an enzyme needed for HSPG biosynthesis and its activity is needed to allow movement of cholesterol-modified Hh-N in A cells
Heparan sulfate (HS) glycosaminoglycan (GAG) chains are synthesized on a core protein by the sequential action of individual glycosyltransferases and modification enzymes, in a three-step process involving chain initiation, polymerization and modification. HS chain synthesis begins with the assembly of a linkage tetrasaccharide on serine residues in the core polypeptide. This process is catalyzed by four enzymes (Xyl transferase, Gal transferase I-II and GlcA transferase I), which add individual sugar residues sequentially to the non-reducing end of the growing chain.
The core protein of Dly is crucially required for Hh signaling, whereas the attached HS chains are required for optimal Hh signaling activity.
1. http://zider.free.fr/papers/Paper%20(13).pdf
2. http://genesdev.cshlp.org/content/15/23/3059.full.pdf
3. http://www.pnas.org/content/103/46/17208.full.pdf
4. INSECT DEVELOPMENT MORPHOGENESIS, MOLTING AND METAMORPHOSIS, page 70
Last edited by Admin on Thu Jun 23, 2016 10:17 am; edited 18 times in total