The Evolution of Early Animal Complexity 1
The argument of the development of an embryo
1a. During the development of an embryo, everything happens at a specific moment. In about 48 hours, it will grow from the top to the bottom, one slice at a time – scientists call this the embryo’s segmentation. “We’re made up of thirty-odd horizontal slices,” explains Denis Duboule, a professor at EPFL and Unige. “These slices correspond more or less to the number of vertebrae we have.”
1b. Every hour and a half, a new segment is built. The genes corresponding to the cervical vertebrae, the thoracic vertebrae, the lumbar vertebrae and the tailbone become activated at exactly the right moment one after another.”
1c. The process is astonishingly simple. In the embryo’s first moments, the Hox genes are dormant, packaged like a spool of wound yarn on the DNA. When the time is right, the strand begins to unwind. When the embryo begins to form the upper levels, the genes encoding the formation of cervical vertebrae come off the spool and become activated. Then it is the thoracic vertebrae’s turn, and so on down to the tailbone. The DNA strand acts a bit like an old-fashioned computer punchcard, delivering specific instructions as it progressively goes through the machine.
1d. “A new gene comes out of the spool every ninety minutes, which corresponds to the time needed for a new layer of the embryo to be built,” explains Duboule. “It takes two days for the strand to completely unwind; this is the same time that’s needed for all the layers of the embryo to be completed.” This system is the first “mechanical” clock ever discovered in genetics; it is so remarkably precise.
1e. The Hox clock is a demonstration of the extraordinary complexity of the species.
2. The scientists don’t offer any evolutionary explanations. By discovering more and more complexities, the God arguments are increasing; we can only explain the complexities as being by God’s creation and control.
3. God exists.
Review: General Pattern of Development
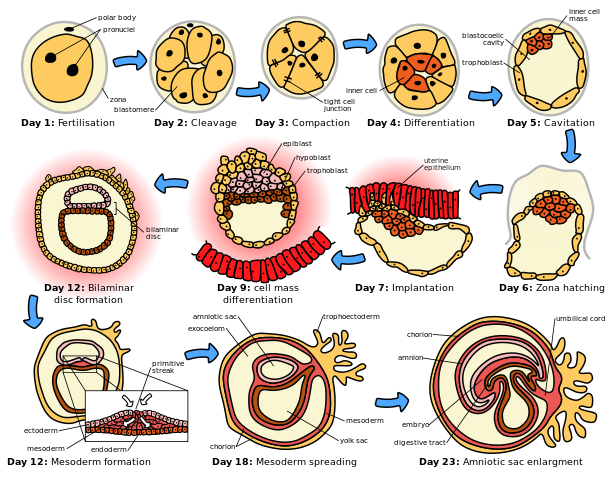
1. Fertilization - In all animals, germ cells produce by meiosis eggs or sperm. The fusion of an egg or sperm to form a zygote is called fertilization. This is the starting point for development.
2. Cleavage - The division of the zygote into smaller and smaller cells.
3. Blastulation - cleavage eventually gives rise to a hollow ball of tiny cells called a blastula.
4. Gastrulation - The sorting out of cells of the blastula into layers (ectoderm, mesoderm, endoderm) that become committed to the formation of future body organs.
5. Differentiation - the formation of body tissues and organs. The basic body plan of the animal is established.
6. Growth - increased size of the animal.
Differentiation is a key feature of multicellular life:
An embryo is a multicellular diploid eukaryote in its earliest stage of development, from the time of fertilization through sexual reproduction until birth, hatching, or germination.
In embryology, cleavage is the division of cells in the early embryo. The zygotes of many species undergo rapid cell cycles with no significant growth, producing a cluster of cells the same size as the original zygote. The different cells derived from cleavage are called blastomeres and form a compact mass called the morula. Cleavage ends with the formation of the blastula.
A morula is distinct from a blastocyst in that a morula (3-4 days post fertilization) is an 8 cell mass in a spherical shape whereas a blastocyst (4-5 days post fertilization) has a cavity inside the zona pellucida along with an inner cell mass. A morula, if untouched and allowed to remain implanted, will eventually develop into a blastocyst.[3]
The morula is produced by a series of cleavage divisions of the early embryo, starting with the single-celled zygote. Once the embryo has divided into 16 cells, it begins to resemble a mulberry, hence the name morula (Latin, morus: mulberry).[4] Within a few days after fertilization, cells on the outer part of the morula become bound tightly together with the formation of desmosomes and gap junctions, becoming nearly indistinguishable. This process is known as compaction.
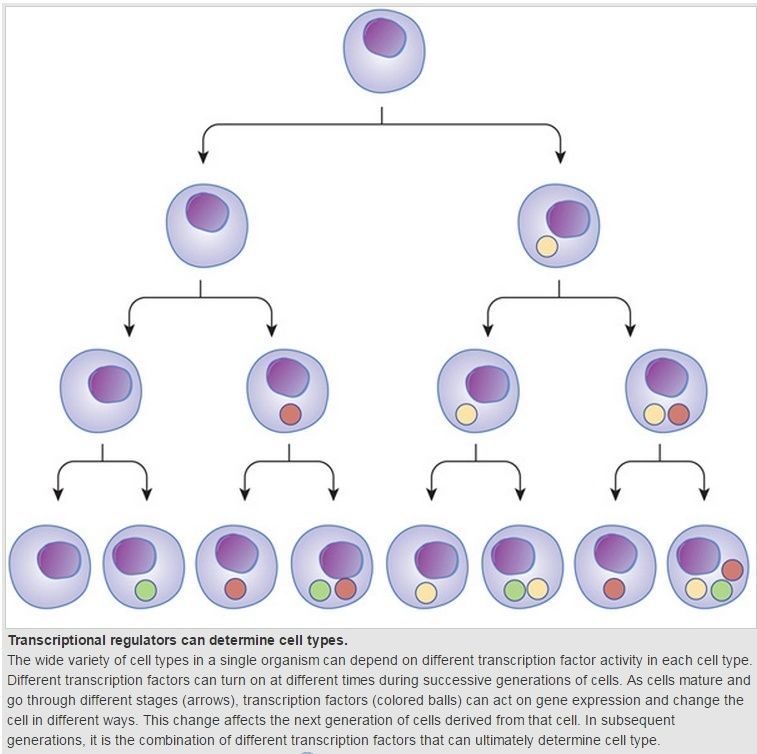
Within the field of developmental biology one goal is to understand how a particular cell (or embryo) develops into the final cell type (or organism), essentially how a cell’s fate is determined. Within an embryo, 4 processes play out at the cellular and tissue level to essentially create the final organism. These processes are cell proliferation, cell specialization, cell interaction and cell movement. Each cell in the embryo receives and gives cues to its neighboring cells and retains a cell memory of its own cell proliferation history. Almost all animals undergo a similar sequence of events duringembryogenesis and have, at least at this developmental stage, the three germ layers and undergo gastrulation. While embryogenesis has been studied for more than a century, it was only recently (the past 15 years or so) that scientists discovered that a basic set of the same proteins and mRNAs are involved in all of embryogenesis. This is one of the reasons that model systems such as the fly (Drosophila melanogaster), the mouse (Muridae), and the leech (Helobdella), can all be used to study embryogenesis and developmental biology relevant to other animals, including humans. What continues to be discovered and investigated is how the basic set of proteins (and mRNAs) are expressed differentially between cells types, temporally and spatially; and whether this is responsible for the vast diversity of organisms produced. This leads to one of the key questions of developmental biology of how is cell fate determined.
1) http://www.gwu.edu/~darwin/BiSc151/NoCoelom/earlyanimal.html
The argument of the development of an embryo
1a. During the development of an embryo, everything happens at a specific moment. In about 48 hours, it will grow from the top to the bottom, one slice at a time – scientists call this the embryo’s segmentation. “We’re made up of thirty-odd horizontal slices,” explains Denis Duboule, a professor at EPFL and Unige. “These slices correspond more or less to the number of vertebrae we have.”
1b. Every hour and a half, a new segment is built. The genes corresponding to the cervical vertebrae, the thoracic vertebrae, the lumbar vertebrae and the tailbone become activated at exactly the right moment one after another.”
1c. The process is astonishingly simple. In the embryo’s first moments, the Hox genes are dormant, packaged like a spool of wound yarn on the DNA. When the time is right, the strand begins to unwind. When the embryo begins to form the upper levels, the genes encoding the formation of cervical vertebrae come off the spool and become activated. Then it is the thoracic vertebrae’s turn, and so on down to the tailbone. The DNA strand acts a bit like an old-fashioned computer punchcard, delivering specific instructions as it progressively goes through the machine.
1d. “A new gene comes out of the spool every ninety minutes, which corresponds to the time needed for a new layer of the embryo to be built,” explains Duboule. “It takes two days for the strand to completely unwind; this is the same time that’s needed for all the layers of the embryo to be completed.” This system is the first “mechanical” clock ever discovered in genetics; it is so remarkably precise.
1e. The Hox clock is a demonstration of the extraordinary complexity of the species.
2. The scientists don’t offer any evolutionary explanations. By discovering more and more complexities, the God arguments are increasing; we can only explain the complexities as being by God’s creation and control.
3. God exists.
Review: General Pattern of Development
1. Fertilization - In all animals, germ cells produce by meiosis eggs or sperm. The fusion of an egg or sperm to form a zygote is called fertilization. This is the starting point for development.
2. Cleavage - The division of the zygote into smaller and smaller cells.
3. Blastulation - cleavage eventually gives rise to a hollow ball of tiny cells called a blastula.
4. Gastrulation - The sorting out of cells of the blastula into layers (ectoderm, mesoderm, endoderm) that become committed to the formation of future body organs.
5. Differentiation - the formation of body tissues and organs. The basic body plan of the animal is established.
6. Growth - increased size of the animal.
Differentiation is a key feature of multicellular life:
An embryo is a multicellular diploid eukaryote in its earliest stage of development, from the time of fertilization through sexual reproduction until birth, hatching, or germination.
In embryology, cleavage is the division of cells in the early embryo. The zygotes of many species undergo rapid cell cycles with no significant growth, producing a cluster of cells the same size as the original zygote. The different cells derived from cleavage are called blastomeres and form a compact mass called the morula. Cleavage ends with the formation of the blastula.
A morula is distinct from a blastocyst in that a morula (3-4 days post fertilization) is an 8 cell mass in a spherical shape whereas a blastocyst (4-5 days post fertilization) has a cavity inside the zona pellucida along with an inner cell mass. A morula, if untouched and allowed to remain implanted, will eventually develop into a blastocyst.[3]
The morula is produced by a series of cleavage divisions of the early embryo, starting with the single-celled zygote. Once the embryo has divided into 16 cells, it begins to resemble a mulberry, hence the name morula (Latin, morus: mulberry).[4] Within a few days after fertilization, cells on the outer part of the morula become bound tightly together with the formation of desmosomes and gap junctions, becoming nearly indistinguishable. This process is known as compaction.

Within the field of developmental biology one goal is to understand how a particular cell (or embryo) develops into the final cell type (or organism), essentially how a cell’s fate is determined. Within an embryo, 4 processes play out at the cellular and tissue level to essentially create the final organism. These processes are cell proliferation, cell specialization, cell interaction and cell movement. Each cell in the embryo receives and gives cues to its neighboring cells and retains a cell memory of its own cell proliferation history. Almost all animals undergo a similar sequence of events duringembryogenesis and have, at least at this developmental stage, the three germ layers and undergo gastrulation. While embryogenesis has been studied for more than a century, it was only recently (the past 15 years or so) that scientists discovered that a basic set of the same proteins and mRNAs are involved in all of embryogenesis. This is one of the reasons that model systems such as the fly (Drosophila melanogaster), the mouse (Muridae), and the leech (Helobdella), can all be used to study embryogenesis and developmental biology relevant to other animals, including humans. What continues to be discovered and investigated is how the basic set of proteins (and mRNAs) are expressed differentially between cells types, temporally and spatially; and whether this is responsible for the vast diversity of organisms produced. This leads to one of the key questions of developmental biology of how is cell fate determined.
1) http://www.gwu.edu/~darwin/BiSc151/NoCoelom/earlyanimal.html
Last edited by Admin on Mon Dec 07, 2015 12:06 pm; edited 2 times in total