Abstract
All extant life forms depend, directly or indirectly, on the autotrophic fixation of the dominant elements of the biosphere: carbon, hydrogen, nitrogen, oxygen, phosphorus, and sulfur. We have earlier presented the canonical network of reactions that constitute the anabolism of a reductive chemoautotroph. Separating this network into subgraphs reveals several empirical generalizations: (1) acetate (acetyl-CoA), pyruvate, phosphoenol pyruvate, oxaloacetate, and 2-oxoglutarate serve as universal starting points for all pathways leading to the universal building blocks—20 amino acids and 4 ribonucleotide triphosphates; (2) all pathways are anabolic; (3) all reactions operate by complete utilization of outputs with no molecules left behind as waste, ensuring conservation of information; (4) the core metabolome of 120 compounds is acidic, consisting of compounds containing phosphoric or carboxylic acid or both; and (5) the core network is both brittle—vulnerable to a single break—and robust—having persisted for 4 billion years. Preliminary analysis of the chemical reactions and resultant structures reveals (a) a sparseness among possible molecular structures; (b) subdomains in the network; and (c) restriction of anabolism to a small set of rudimentary organic reactions with limited diversity in chemical mechanisms. These generalizations have implications for biogenesis and trophic ecology.Introduction
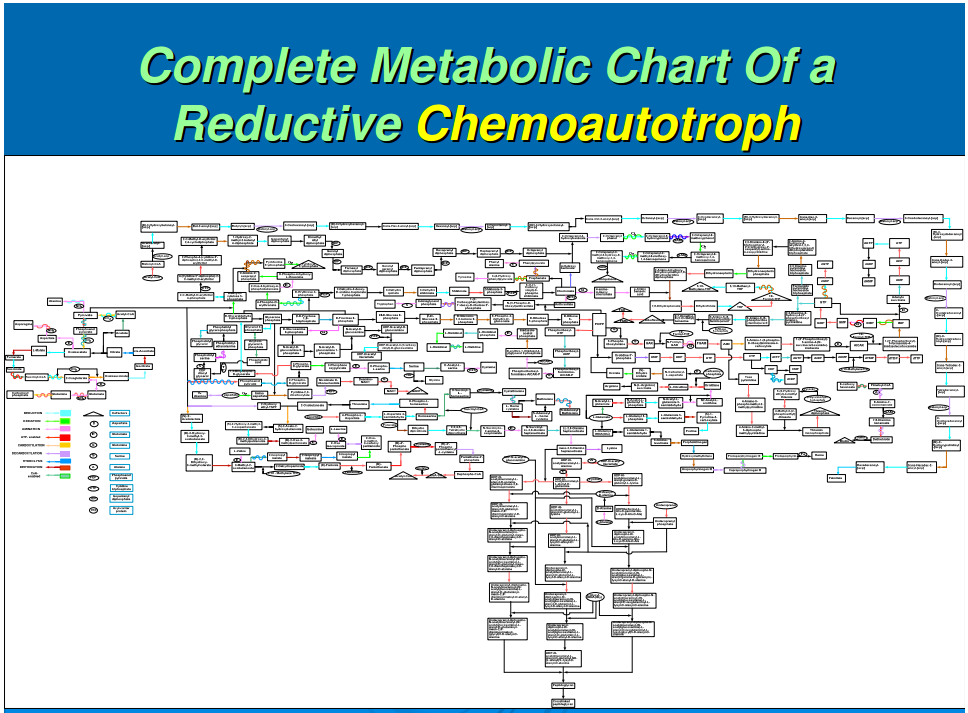
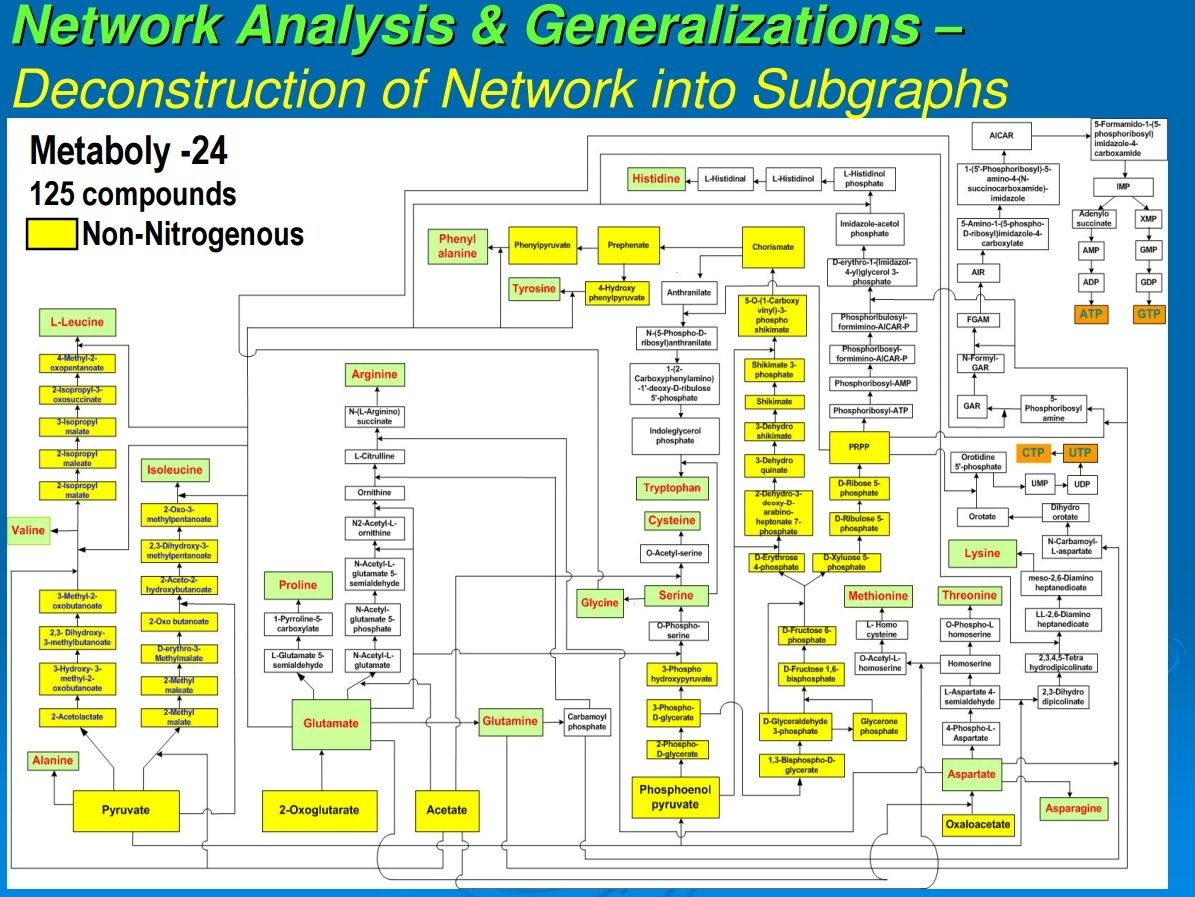
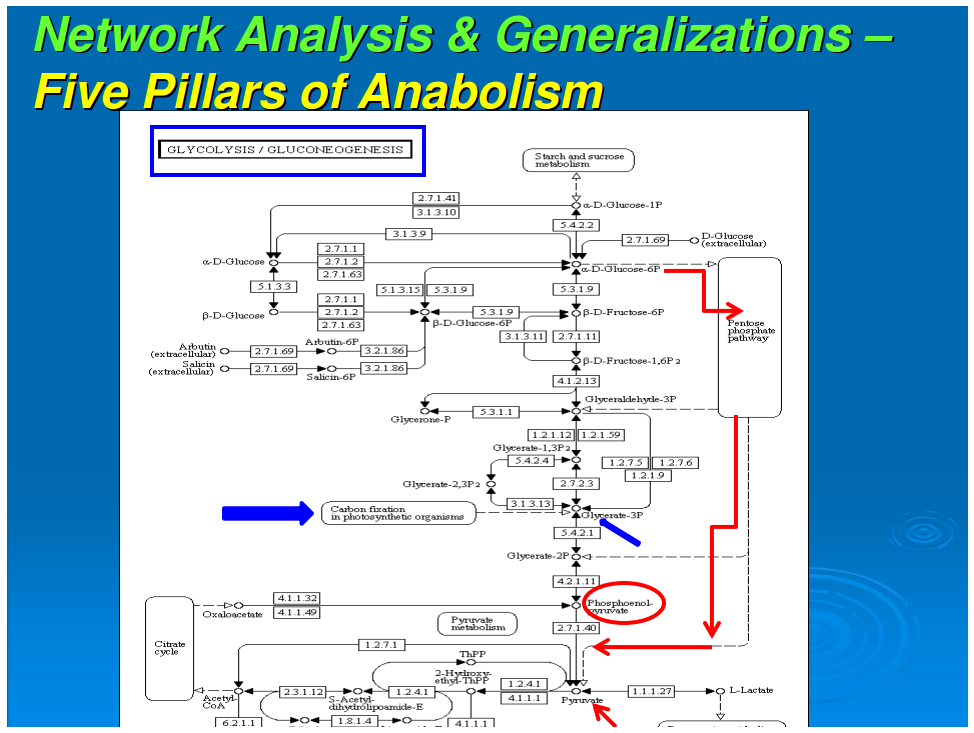
In our previous work, we had data-mined genomic databases and utilized available biochemical data on metabolic pathways of several reductive chemoautotrophic organisms that included Aquifex aeolicus, Hydrogenobacter thermophilus,Thiomicrospira denitrificans, and Chlorobium terpidium, enabling us to construct an almost complete chart of reductive autotrophic intermediary metabolism (Srinivasan and Morowitz, 2009). This metabolic chart of 286 reactions yielding 287 unique compounds constitutes the minimal metabolome of a reductive autotroph. We had broadly classified these reactions into a few types of rudimentary organic chemical reactions. The compounds were classified into two main categories—monomers and intermediates—and other subcategories. From a network perspective, these compounds of the metabolome were also classified into subgroups depending on their nodal status. We now proceed to extend the analysis to other taxa and to deconstruct that map into a group of subnetworks. From these components we then extract a series of empirical generalizations that may provide some insight into biogenesis and general laws of anabolic biochemistry and ecology. The subnetworks are (1) the core metabolism going from the molecules of the five starting points of autotrophic anabolism to the 20 coded amino acids and 4 ribonucleotides that would have existed in the RNA world, (2) the pathways leading from core metabolism to the standard cofactors or coenzymes of extant biochemistry, (3) the pathways from acetate to the polar lipids that become the cell membrane components, (4) the isoprenoid pathways leading to ubiquinone and hopanoids, and (5) the peptidoglygan synthesis pathways. The first two subnetworks that we address in this paper are shown in Figures 1–3. Note that we are focusing on the anabolic networks and assume the presence of energy sources in the form of pyrophosphates and redox couples in these systems.Autotrophic metabolic network of the core metabolome. Complete pathway connectivities and reactions are shown in lines and arrows, respectively. Each box represents a unique compound of the metabolome. The 20 amino acids (green boxes) and 4 ribonucleotide triphosphates (orange boxes) constitute the basic building blocks. Boxes colored in yellow represent compounds prior to the entry and integration of nitrogen to C, H, O, P, and S.
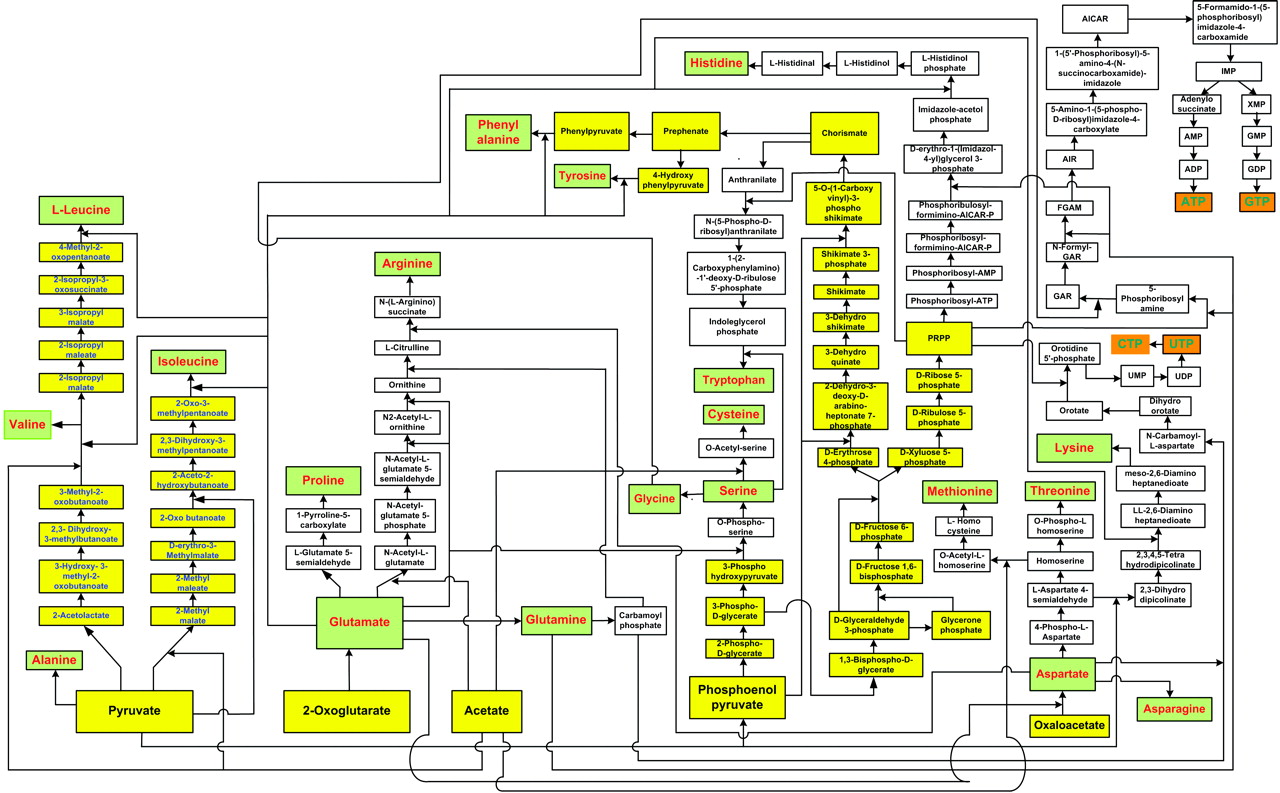
Autotrophic metabolic network of the core metabolome. Complete pathway connectivities and reactions are shown in lines and arrows, respectively. Each box represents a unique compound of the metabolome. The 20 amino acids (green boxes) and 4 ribonucleotide triphosphates (orange boxes) constitute the basic building blocks. Boxes colored in yellow represent compounds prior to the entry and integration of nitrogen to C, H, O, P, and S.
Materials and Methods
All information regarding the pathways, the enzymes, the reactants, and the products for all the organisms whose genomes have been completely sequenced was obtained by data-mining the Kyoto Encyclopedia of Genes and Genomes (KEGG) database—an online, public domain, open-source collection of databases that includes metabolic pathways, genome sequences, and analysis (Kanehisa and Goto, 2000). The KEGG Pathway Database was utilized for retrieval of all information, using the search criteria related to “organisms,” “gene catalogs,” “KEGG Orthology” under “network hierarchy” and “metabolism,” and the entries for each of the enzymes mentioned therein. We have extracted pathway information for the anabolic synthesis of the compounds of the autotrophic metabolome from the KEGG PATHWAY database. During reconstruction, in the few instances we encountered gaps, we adopted the strategy of comparative analysis that has been detailed in our previous work (Srinivasan and Morowitz, 2009) to complete the biosynthetic pathways and the network.Results and Discussion
The empirical generalizations applicable to the entire metabolic map detailed in the previous paper (Srinivasan and Morowitz, 2009) are as follows:
The universal atomic constituents of metabolism are carbon, hydrogen, oxygen, nitrogen, phosphorus, and sulfur.
All autotrophic pathways are anabolic.
When an anabolic pathway results in the splitting of a molecule, all of the resulting product molecules enter into other anabolic pathways. We have designated this phenomenon as “No molecule left behind.” This generalization involves a conservation of information as well as a conservation of inputs in autotrophic anabolism.
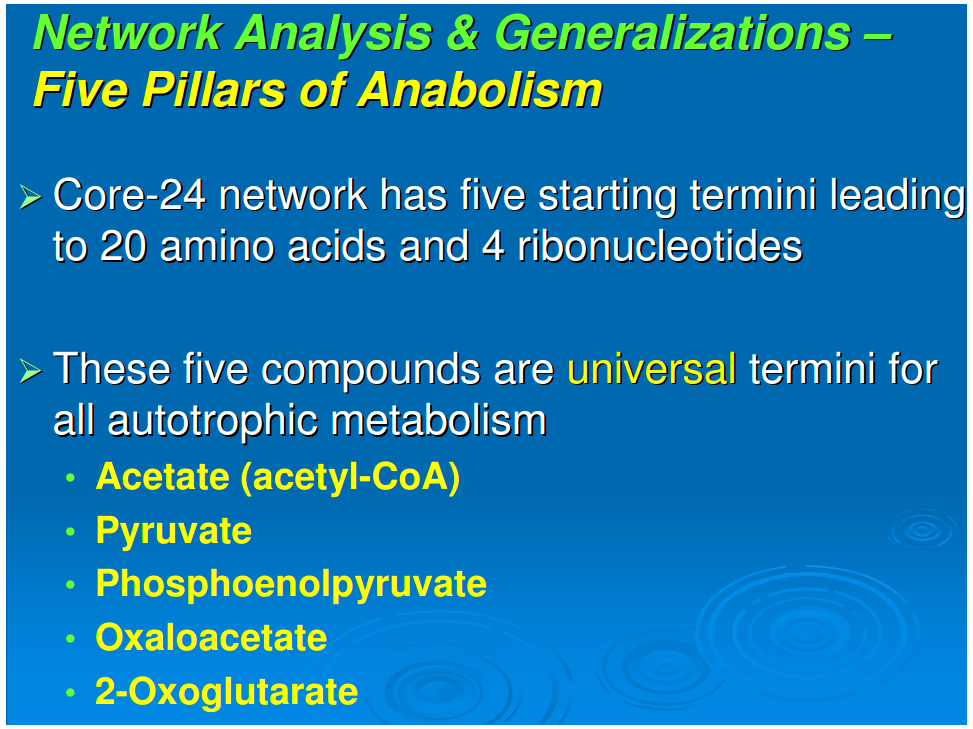
[*]All anabolic networks have their starting points for all of the synthetic pathways in the following five nodal molecules: acetate (acetyl-CoA), pyruvate, phosphoenol pyruvate, 2-oxoglutarate, and oxaloacetate. This generalization appears to be universal for all autotrophs, including those that use the reductive TCA cycle (Fuchs, 1989), those that employ the oxidative TCA cycle (Nelson and Cox, 2000), and those with the “horseshoe,” or incomplete, TCA cycle (Ljungdahl, 1986; Ragsdale, 1991). This will be discussed in detail below.
We next focus attention on the core metabolism shown in Figure 1. This is the network for all anabolic synthesis and it is the subgraph of the pathways for the basic building blocks in the main metabolic chart presented earlier for reductive chemoautotrophs (Srinivasan and Morowitz, 2009); it is presumably universal for all autotrophs. As such it is the core network for ecological biochemistry as well as for autotrophic organisms since all large-scale ecological systems go through an autotrophic primary trophic level. At this stage, three more generalizations enter for the core pathway:
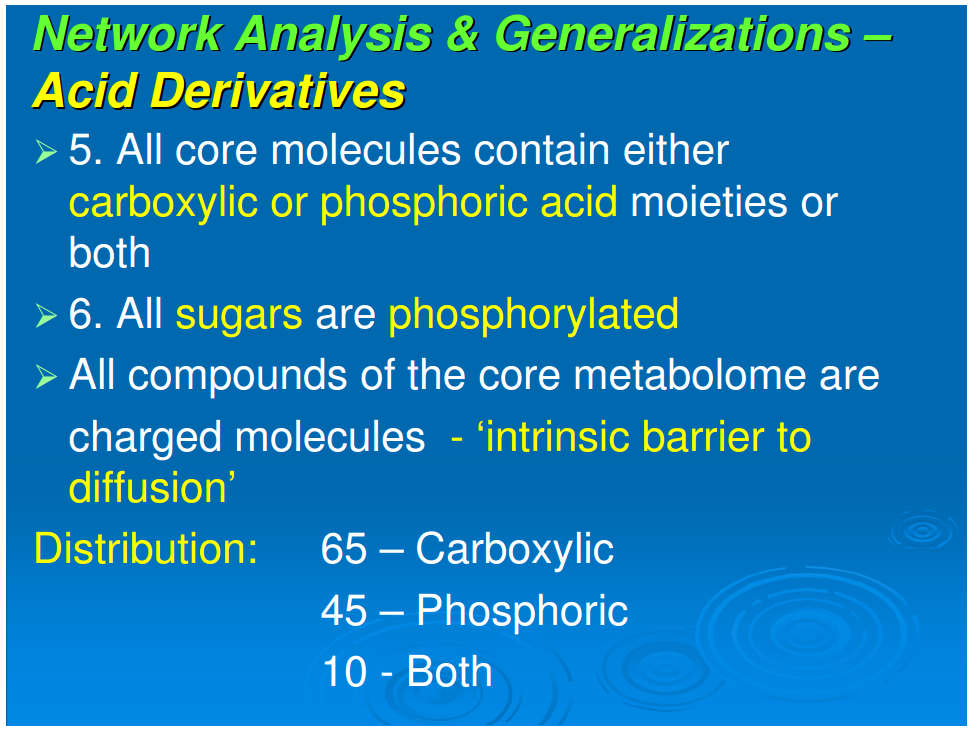
5. All sugars are phosphorylated.
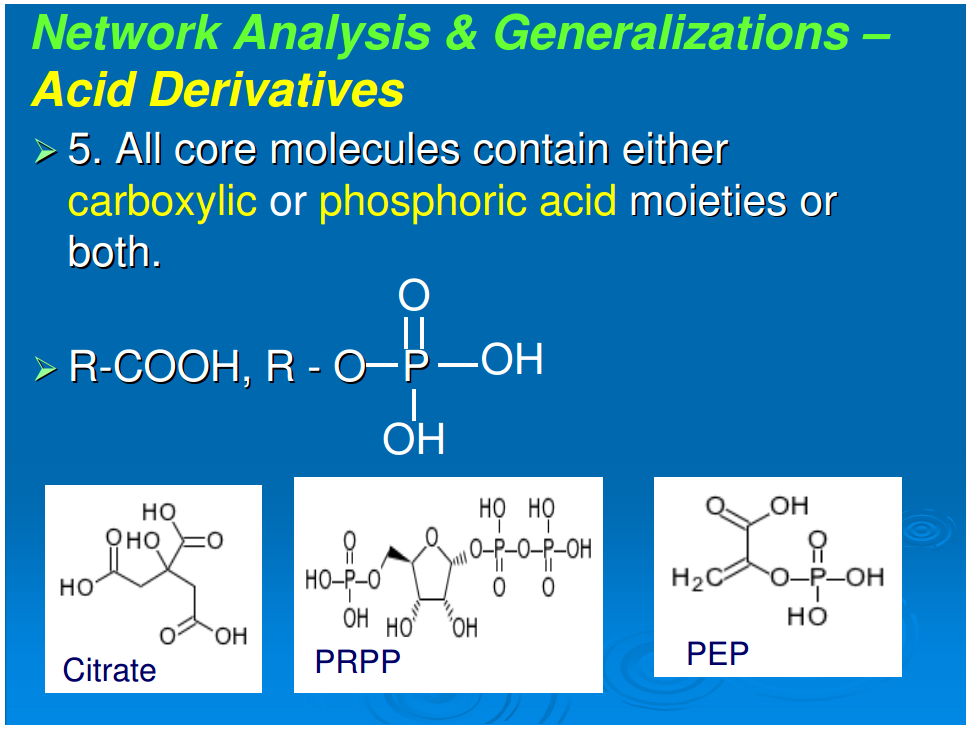
6. All core molecules contain either phosphoric or carboxylic acid moieties or both. The possible exceptions, histidinal and histinidol, may have resulted from components of a vestigial cofactor pathway.
7. The core anabolic network is brittle in the sense that a break in any link undermines the complete availability of the basic building blocks (20 amino acids and 4 ribonucleotides) necessary for the synthesis of macromolecules and for the buildup of the next hierarchical level.
The 125 compounds of core biochemistry shown in Figure 1 are a strikingly sparse subset of the domain of small molecules that may be made of C, H, N, O, P, and S, conformed to the observed atomic composition range for each element in the complete set (Fink et al., 2005). Core biochemistry is remarkably robust as well as being brittle, having been extant for some 4 billion years and having persisted in spite of vast changes in the array of enzymes and their genetic embodiments; that is, the core biochemical network is more stable than the rich variety of enabling molecules that reify it across the taxa.
[*]
Generalization 1: C, H, N, O, P, S— the universal atomic components
Almost 99% of the metabolically active components of living organisms is composed of carbon, hydrogen, nitrogen, and oxygen, and the rest is made up largely of phosphorus and sulfur. The explanation of the atomic composition of biota was discussed in 1961 in an extraordinarily perceptive and fundamental paper by Nobel Laureate George Wald entitled “Life in the Second and Third Periods; or Why Phosphorus and Sulfur for High Energy Bonds” (Wald, 1962). The choice of C, H, N, and O can be readily attributed to their smallness, which confers the ability to form multiple and stable bonds. Although carbon, the basis of all biological material, has a relative abundance of only 0.119% compared to silicon's 16.08%, the relative instability of Si–Si bonds in the presence of water, ammonia, or oxygen precludes choice of silicon as the major element of life. Phosphorus in the presence of water is almost always orthophosphate or pyrophosphate, and the critical attributes for its incorporation into biochemicals are its thermodynamic instability combined with kinetic stability, its charge, and a constant oxidation state under biological redox conditions. Similarly, the presence of sulfur as a key element in biology is due to its ability to form significantly weaker bonds than its congeners oxygen and nitrogen. This reduced stability in bonding confers a ready ability to combine with other molecules and thus promotes facile group transfer and exchange reactions.
Generalization 2: All pathways are anabolic
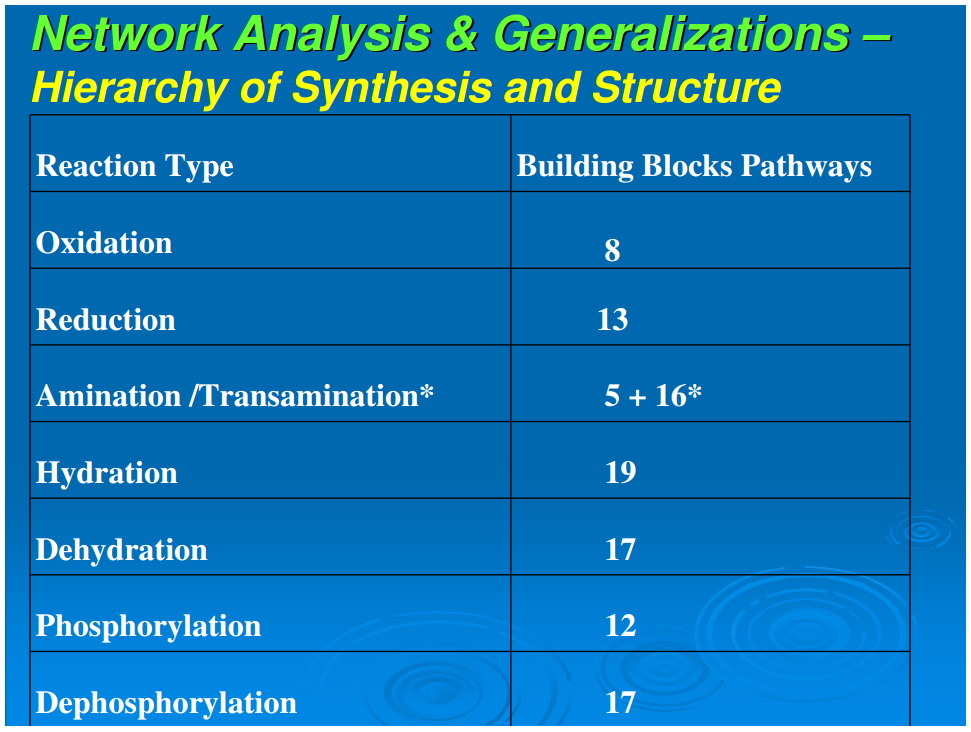
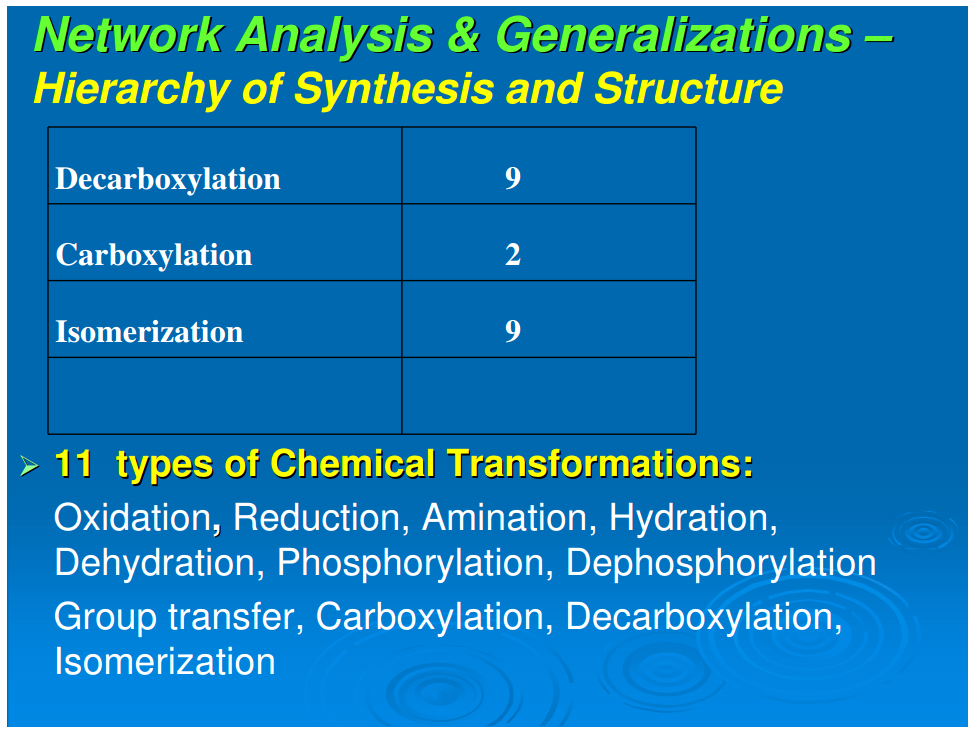
A simplifying feature of all reductive autotrophs, which constitutes a major distinction from heterotrophs, is that there are no catabolic pathways in the former. This appreciably simplifies the metabolic maps of reductive autotrophs and allows for the next generalization: that all carbon fixation pathways, oxidative and reductive, yield outputs that are routed through gluconeogenesis and glycolysis (catabolic pathways that occur only in the oxidative heterotrophs), and oxidative or reductive TCA cycles, or parts of these cycles.In autotrophic anabolism, the source materials are developed in discrete chemical reactions in defined pathways to produce the building blocks of all cellular material. It is striking that the types of organic reactions in core anabolism are limited to 11 chemical transformations: hydrolysis/dehydration, carboxylation/decarboxylation, oxidation/reduction, phosphorylation/dephosphorylation, transamination, group transfer, and isomerization (Table 1). In the known repertoire of reaction types in organic synthesis, this is a relatively very small set. From the point of view of biogenesis, it is noteworthy that the requirements of the types of organic reactions necessary for the emergence of this organized core metabolic network are surprisingly rudimentary
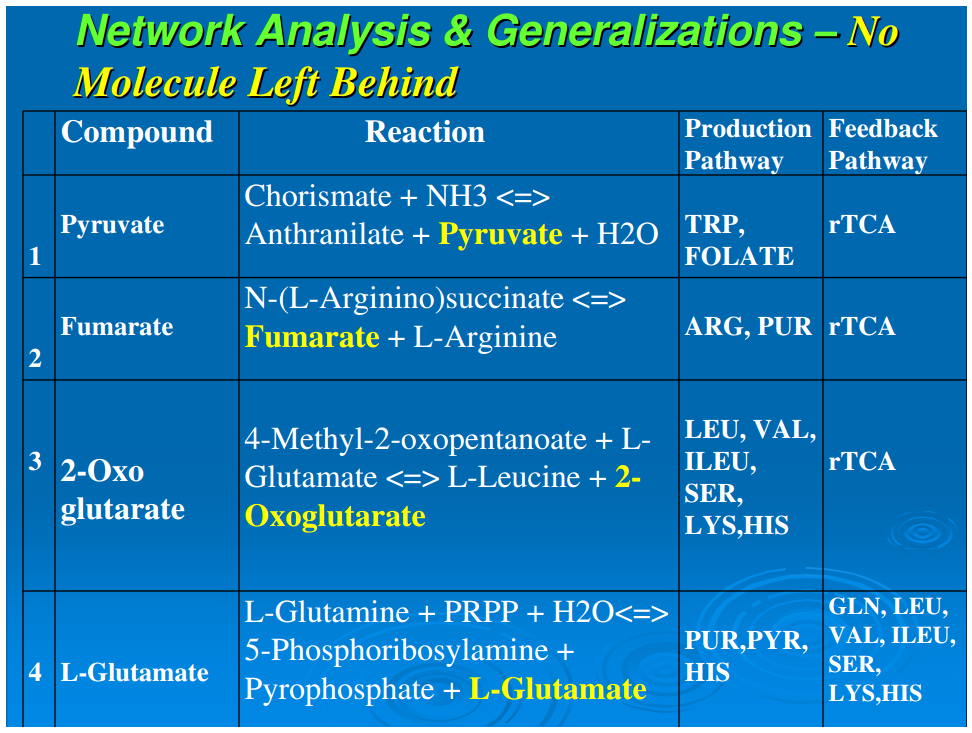
Generalization 3: No molecule left behind
In heterotrophic biochemistry, all pathway reactions generate product molecules that enter into anabolic pathways, enter into catabolic pathways, or are secreted. This generalization is that in autotrophic metabolism, all products go into anabolic pathways, with all inputs completely utilized, and therefore generate no waste products.Table 2 depicts representative examples of reactions that involve simple molecules to compounds of relatively higher complexity like AICAR, and it is evident that all the product molecules are entirely utilized—the main product progresses to the next step and the rest of the products are fed back into the network for other reactions. Thus if a molecule splits and one of the products is used in one anabolic pathway, the other product is always used in another anabolic pathway. This is a highly efficient mode of operation in that not only are no molecules wasted, but no synthetic organic reactions are wasted. This mechanism enforces a conservation of information in autotrophic anabolism, and we have designated this feature as “No Molecule Left Behind.” The observed biofrugality appears to be an effective microeconomic principle in which all outputs (synthesized material) are completely utilized or recycled into the core network for processing. Therefore, systems with this kind of efficiency have a competitive advantage at a deep biochemical level, and this offers another way to analyze ecosystems from a biochemical network perspective. We believe that the “no molecule left behind” principle for reductive autotrophs is a deep general biological principle that should be examined in other autotrophic taxa for ecological significance.
Generalization 4: Five pillars of anabolism
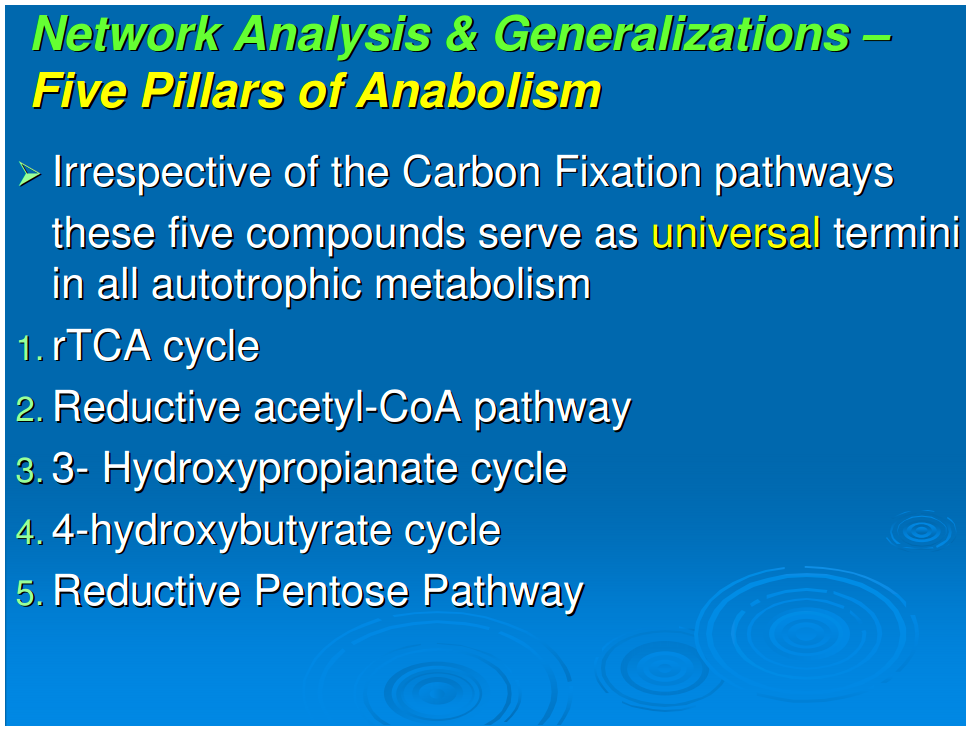
Figure 1 shows the pathways for a reductive autotroph in which the five starting points generated in the reductive TCA cycle lead stepwise to the canonical 20 amino acids and 4 ribonucleotides. In comparing the anabolic pathways of other autotrophs, which include phototrophs, we observed a universality of this feature of anabolism. The universal starting points are acetate (acetyl-CoA), pyruvate, phosphoenol pyruvate, oxaloacetate, and 2-oxoglutarate. At the present time five different autotrophic pathways have been described (Thauer, 2007; Nakagawa and Takai, 2008) and we chose these for comparison, all having been well established for carbon assimilation: (i) the reductive pentose phosphate pathway (Calvin and Bassham, 1962), (ii) the reductive tricarboxylic acid (rTCA) cycle (Buchanan and Arnon, 1990), (iii) the reductive acetyl-CoA pathway (Ljungdahl, 1986), and (iv) the 3-hydroxypropianate cycle (Ishii et al., 2004) and (v) its variant the 4-hydroxy butyrate cycle (Berg et al., 2007).In anaerobic autotrophic bacteria, carbon fixation is accomplished by pathways that share the reductive synthesis of the common intermediate, acetyl-CoA, generated from 2CO2 and 8[H] (Fuchs, 1989). The reductive acetyl-CoA pathway is characterized by the presence of the key enzyme complex acetyl CoA synthetase, which also catalyzes the oxidation of CO to CO2. Also known as the Wood-Ljungdahl pathway, it is a non-cyclic route that synthesizes acetyl-CoA as the key intermediate and is mostly functional in archaea, especially the acetogens and methanogens.The reductive tricarboxylic acid (rTCA) pathway runs in the opposite directions from the well-known Kreb's cycle—the oxidative citric acid cycle (Nelson and Cox, 2000). The marker enzymes of this pathway are the ATP-citrate lyase and the 2-oxoglutarate synthetase, which characterize the productions of the key intermediates acetyl-CoA, oxaloacetate, and 2-oxoglutarate (Hügler et al., 2005).The 3-hydroxypropianate cycle is another unidirectional pathway in which acetyl-CoA is reductively transformed through carboxylation reactions into succinyl-CoA via the 3-hydroxypropianate intermediate. Succinyl-CoA is converted to acetyl-CoA that is fed back into the cycle, with the net result of generating glyoxylate as the carbon fixation product, which is converted to pyruvate (Herter et al., 2002). Malonyl-CoA reductase and propionyl-CoA synthetase are the key enzymes of the 3-hydroxypropianate cycle, which is operative in Chloroflexus (Ishii et al., 2004) with a modified version detected in the crenarchaeal order Sulfolobales (Alber et al., 2006).Recently, a 3-hydroxypropianate/4-hydroxybutyrate cycle has been uncovered as another pathway for autotrophic carbon fixation (Berg et al., 2007). Here, an acetyl-CoA molecule combines with two carbon dioxide units and gets reductively converted, first to 3-hydroxypropianate and then to 4-hydroxy butyrate, eventually yielding two acetyl CoA molecules in a reaction catalyzed by 4-hydroxybutyryl-CoA dehydratase. Discovered first in autotrophic members of the archaeal order Sulfalobus, the key genes for this pathway have been detected in Sulfalobus, Archaeoglobus, and Cenarchaeumspecies and are believed to be very widespread, as inferred from the Global Ocean Sampling Database (Berg et al., 2007). In all of these anaerobic carbon fixers, the five starting compounds for anabolism appear to be the same.In carbon fixation by aerobes, the reductive pentose phosphate pathway is functional in chloroplasts and utilized by most plants (Calvin and Bassham, 1962). Ribulose 1,5-bisphosphate carboxylase/oxygenase (RUBISCO) and ribulose 5-phosphate kinase (phosphoribulokinase) are the marker enzymes of this pathway, and the cycle runs unidirectionally to produce 3-phosphoglycerate as a key intermediate. Also known as the Calvin-Bassham-Benson cycle, this pathway operates in cyanobacteria, the progenitor of chloroplasts, as well as in plants and most aerobic and facultative anaerobic autotrophic bacteria. The synthesized sugars lead to the oxidative citric acid cycle with the anaplerotic loop. For such aerobic autotrophs, we should add 3-phosphoglycerate to the five starting points because the anabolic pathways go through gluconeogenesis and a catabolic route to the citric acid cycle and then the anabolic pathways to the synthesis of amino acids and ribonucleotides. There is thus a universal anabolic map for all autotrophs. It applies to both chemo- and photo-autotrophs and is independent of their carbon fixation strategy—including those that use the reductive TCA cycle, those that use the oxidative TCA cycle, and those with the “horseshoe,” or incomplete, TCA cycle. Since ecosystems are “in toto” autotrophic, this anabolism is a general feature of ecology.There are a limited number of sites along the anabolic pathways where carbon is further incorporated directly from CO2. The first of these is the synthesis of carbamoyl phosphate, which is in a pathway originating in 2-oxoglutarate. A second site is the carboxylation of aminoimidazole ribotide in purine synthesis. The synthesis of fatty acids involves the repetitive addition of CO2 to synthesize malonic acid for chain extension in fatty acid production.
Generalization 5: All sugars are phosphorylated
This generalization applies to the core anabolic network shown in Figure 1. Sugars are involved in the synthesis of two nodal intermediates, chorismate and PRPP. Chorismate is required for the synthesis of the aromatic amino acids phenylalanine, tyrosine, and tryptophan; PRPP is involved in the synthesis of histidine and also provides the ribose component for the purine and pyrimidine nucleotides. The sugars are universally phosphorylated, both individually and in the compounds in which they are structurally integrated into the components of core metabolism, as seen in Figure 1. Phosphorylation appears to impart further charge to the sugar molecules that are polar (by virtue of carrying −OH hydroxyl groups), and this further enhances their solubility as well as reinforcing the barrier to diffusion outside. Furthermore, that all sugars are phosphorylated is consistent with our observation that all compounds of the core metabolome are acid derivatives (see below).
Generalization 6: All core molecules contain either phosphoric or carboxylic acids or both
Referring to Figure 1, all compounds are derivatives of carboxylic or phosphoric acid or both, with the exception of histidinol and histidinal. These exceptions, as noted above, may be part of a cofactor pathway that was incorporated into an amino acid in order to place the imidazole functional group in proteins (unpubl. data). Excluding these two compounds, 65 of the core metabolome acids contain carboxylic acid, 43 contain phosphoric acid, and the remaining 10 contain both acid groups. The acidic character of almost all of the intermediates in monomer synthesis is a generalization that must be of some significance. At neutral pH, all the intermediates must be negatively charged. From the viewpoint of biogenesis, it is possible that this attribute facilitated maximizing the concentration of the core metabolic components by preventing their diffusion through nonpolar, hydrophobic encapsulations or by stabilizing their adsorption on charged surfaces.
Generalization 7: The core anabolic network is both brittle and robust
The core network is both sparse and non-redundant and represents a highly efficient way of producing the 24 essential compounds. As a result, any break in the network ofFigure 1 results in an inability to make one or more of the required molecules for macromolecular synthesis. This feature is described as brittleness. On the other hand, the ubiquity of the network and its persistence for 4 billion years and through vast evolutionary variation in enzymatic sequence and performance suggest a robustness in the network that supersedes the individual hardware. We have referred to this as selfish metabolism (Morowitz et al., 2009). Thus we observe the enigma of brittleness and robustness. A related study pertaining to heterotrophs (Samal et al., 2006) presents another example of brittleness in the context of modular organization.
Assimilation of C, H, N, O, P, and S
Autotrophic carbon fixation by the available pathways has been discussed in the preceding paragraphs. While nitrogen exists in nature in a wide variety of redox states from nitrate to ammonia, its entry into anabolic metabolism is almost universally in its most reduced form, NH3. As a consequence, autotrophs and ecosystems must have various nitrogen reductase systems leading to ammonia. Its assimilation is then enabled by the coupled action of the enzymes glutamate synthetase (glutamine: 2 oxoglutarate amidotransferase [GOGAT]) and glutamine synthetase (GS) (Kameya et al., 2006,2007). This GOGAT/GS pathway operates widely across all three taxa: bacteria, archaea, and eukarya. Glutamine synthase incorporates NH3 into glutamate to produce glutamine, which in a coupled reaction, catalyzed by glutamate synthase, combines with 2-oxoglutarate to generate two molecules of glutamate. Subsequently, this glutamate serves as a nitrogen donor to be distributed into the monomers and multimers predominantly through transamination reactions enabled by the cofactor pyridoxal phosphate. This seems like an ecologically universal way for nitrogen to enter into anabolic metabolism and thus constitutes an ecological principle. The major sources of oxygen for autotrophic assimilation are CO2, H2O, and phosphate. The origin of phosphorus for the compounds of the autotroph metabolome can be traced mostly to ATP as well as to related nucleoside phosphate derivates. From the viewpoint of biogenesis, phosphoric acid and polyphosphates may have served as the prebiotic source for phosphorylation reactions. Sulfur also seems to enter into anabolism in its most reduced form, H2S. It first gets integrated into cysteine, which upon conversion to homocysteine leads to methionine and S-adenosyl methionine.
Autotrophic mode of entry for carbon, hydrogen, nitrogen, oxygen, phosphorus, and sulfur
Carbon enters in multiple states: the most reduced is the methyl (CH3); the intermediate states of oxidation are the hydroxymethyl (CH2–OH), aldehyde (H–C=O), and carboxylic groups (OH–C=O); and the most oxidized form for incorporation is carbon dioxide (O=C=O). Entry of hydrogen occurs via H–H or complex redox chemistry of cofactors, and nitrogen enters as NH3 and gets incorporated into glutamate and glutamine first. Hydroxyl(–OH) and carbonyl(C=O) groups are the modes of entry for oxygen, phosphorus enters as phosphate that is mostly stabilized with the metal ion magnesium, and sulfur's entry is limited to H2S that gets incorporated into cysteine. It is interesting that for each of the different modes of entry of C, H, N, O, P, and S, there is at least one cofactor present that enables their incorporation into components of the metabolome (unpubl. data).
Hierarchy of synthesis and structure
The core anabolic network shown in Figure 1 consists of a series of chemical reactions building up compounds step by step. It defines elementary anabolism, and the reaction types involved are presented in Table 1. Not shown in this summarization are the catalytic effects that some molecules in this collection have on the reactions of the network. The first level of synthesis, akin to normal organic synthesis, seems to operate up to a molecular weight of 200 to 300 Da, which defines the range of small molecules that are pathway intermediates and monomers. The hierarchical order of synthesis produces the following:[list=list-ord]
MonomersPolymersChimeromersRepeatomersSuper chimeromers such as peptidoglycanCoacervates and other structures held together by noncovalent bonds[/list]
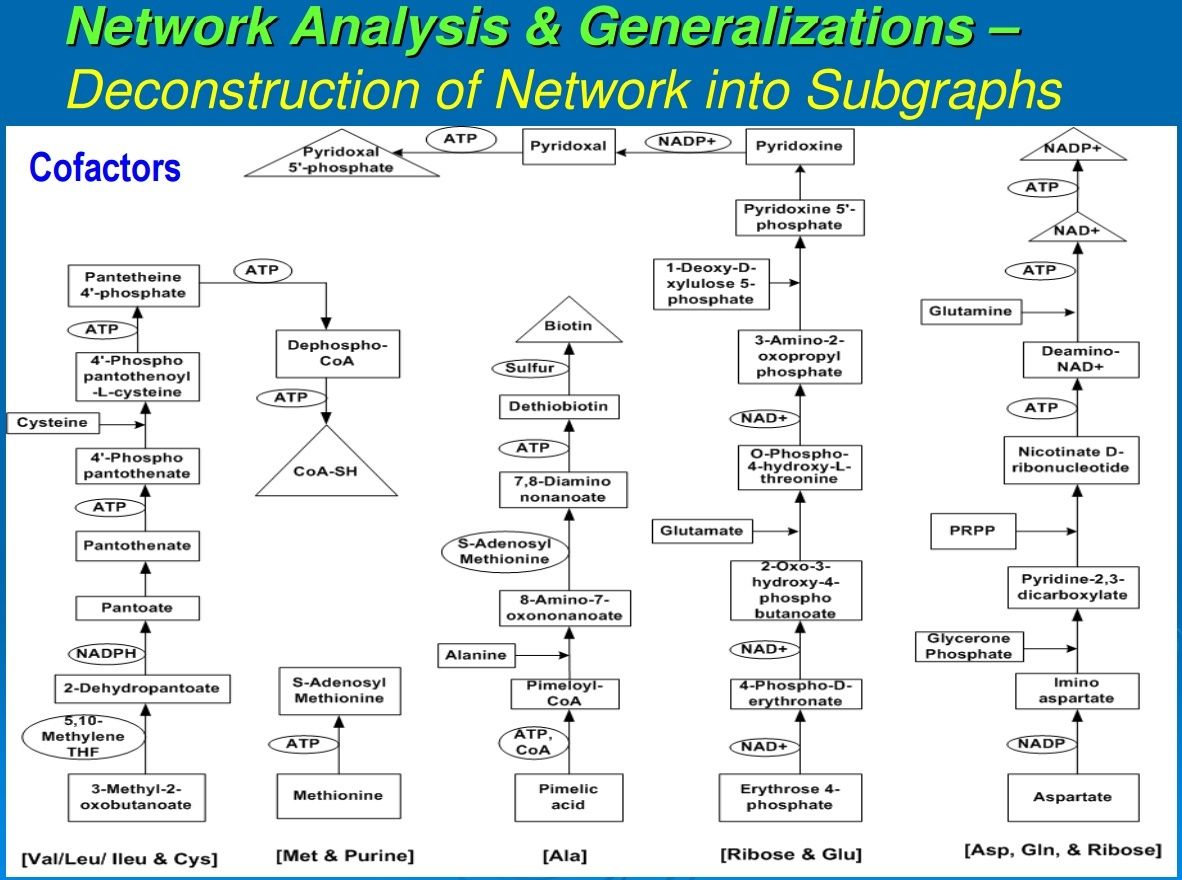
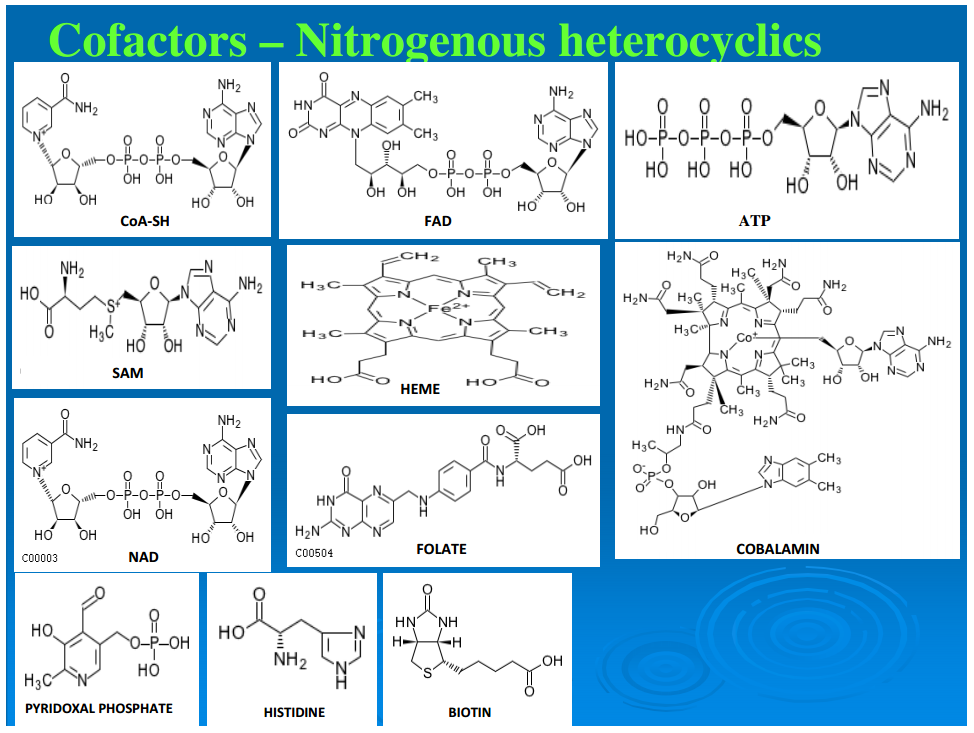
Figure 2 illustrates a representative example for each of these classes of compounds. A second order of synthesis involves forming a covalent bond between two molecules, extracting an OH from one and an H from the other. This dehydration reaction is generally driven by the hydrolysis of a pyrophosphate bond. If the two molecules or higher aggregates have the same backbone structure, the molecules thus formed are called polymers. This may lead to chains of hundreds or more of monomer units, and proteins and nucleic acids are typical examples. If molecules of different monomers and pathway intermediates combine, the resultant structures are designated as chimeromers, and they are generally only a couple of monomers in length. Examples are coenzyme A and riboflavin. Coenzymes are most often formed from chimeromers. Yet another type of product of synthesis we designate as “repeatomer.” Examples are heme and a collection of closely related biological porphyrins. Succinate and glycine condense to form aminolevulinate, and two molecules of this intermediate condense to form porphobilinogen. Four porphobilinogens condense to form a ring of repeatomer of which heme is an example. Super chimeromers such as peptidoglycan can be made of units of two amino sugars and a string of amino acids. They can then be cross-linked at three or four points to form a gigantic structure—the cell wall with a molecular weight up to millions of daltons. The next level consists of structures held together by noncovalent bonds. These aggregates include the ribosomes, the early coacervates, and the extant cell membranes.Subgraph of pathways leading to biosynthesis of cofactors (Group A) of reductive autotrophic metabolism. Shown in brackets, at the bottom, are the names of the individual pathways from which compounds are utilized, mostly in condensation reactions to form chimeromers. Arrows show chemical transformations, and each box represents a unique compound showing synthetic progression from start compounds (shown in boxes at the base) recruited from the core metabolome. Cofactors enabling other cofactors’ biosynthesis are shown in circles; cofactors shown in triangles are the terminal products in their synthesis.
Most of the reactions in the canonical anabolic chart require enzymes that may be isolated apoenzymes or holoenzymes (a combination of apoenzymes and coenzymes and/or metal ions). Figures 3 and 4 trace the synthetic pathways from the core anabolic chart to the coenzymes of a representative chemoautotroph (Srinivasan and Morowitz, 2009). From a biogenesis perspective, contemporary coenzymes appear to have emerged by the fine-tuning of simpler structures. For example, ATP as a cofactor essentially functions as a polyphosphate; and CoA, although a molecule of several hundred daltons, operates as R–SH, the functional part being just the sulfhydryl group, SH. Also, as apparent from Figures 3 and 4, in the extant pathways of cofactor biosynthesis, each cofactor requires the involvement of its own cofactors and/or one or more other cofactors. This further supports the contention that some vestigial form of the current cofactors must have existed before the fine-tuning of their structures and the evolution of their biosynthetic pathways (Begley, 2006). It seems likely that in the earliest biochemistry, simpler molecules that are nonpolymers carried out the role of the cofactors, which now form a small set of sophisticated chemical operators, functioning in conjunction with polypeptides. Since they are all made from molecules in the canonical anabolic chart, they can be traced back to the five anabolic points. Protein catalysts then consist of a polypeptide with the possible addition of coenzymes and metal ions. There is a set of about 10 cofactors that seem to be universal across the taxa. An additional 9 cofactors are found in methanogens and acetogens (Ferry and Kastead, 2007).Subgraph of pathways leading to biosynthesis of cofactors (Group B) of reductive autotrophic metabolism. Shown in brackets, at the bottom, are the names of the individual pathways from which compounds are utilized, mostly in condensation reactions to form chimeromers. Arrows show chemical transformations, and each box represents a unique compound showing synthetic progression from start compounds (shown in boxes at the base) recruited from the core metabolome. Cofactors enabling other cofactors’ biosynthesis are shown in circles; cofactors shown in triangles are the terminal products in their synthesis.
Structural hierarchy in autotrophic metabolome. The symbol “]n” in the Polymer diagram indicates further extension of the chain at both the NH–and CO–links with more amino acids. In the Superchimeromer diagram, ‘R’ represents amino acid side chain. The Aggregate diagram is a schematic representation of cell membrane in which proteins are noncovalently embedded in the lipid bilayer.
General features
A global analysis of the autotrophic network of Figure 1 indicates two other significant general features: (1) the sparseness of the complete set comprising pathway intermediates and the starting point compounds; (2) the restricted set of chemical mechanisms employed in going from the 5 terminal molecules to the set of 24.Sparseness.
An estimation of the possible number of covalently bonded compounds of CHNOPS with a molecular weight less than 300 Da, conforming to the maximal size of the core components, yields a number at least in the millions and probably several orders of magnitude higher (Fink et al., 2005). The actual number of compounds in core metabolism is 125. The actual set is thus a strikingly small subset of the possible. This means that the pruning rules, whatever they may be, generate a very restricted set of compounds. Thus, radical sparseness is clearly a very important feature of canonical autotrophic metabolism. The combination of the observed sparseness, ubiquity, brittleness, and robustness of the core metabolic network suggests a lawfulness to core biochemistry deep within physical and geochemical domains.Chemical mechanism.
We next examine Figure 1 to analyze the set of reaction types necessary to generate the 24 building blocks from the five starting points. Table 1 summarizes the distribution of reaction types in their biosynthetic pathways to the building blocks. It is seen that a limited set of simple reactions are recursively used to perform the progression from source molecules all the way to the terminal products. To place this in context, we note the statement by Petsko and Dagmar (2004) that “about three quarter of the reactions of metabolism can be described by as few as four general types of chemical transformations: oxidation/reduction; addition/eliminations; hydrolysis, and decarboxylation.” We make a somewhat finer division, and that expanded version is shown in Table 1. Nevertheless, these reactions are basic, well-understood mechanisms of organic chemistry. The sparseness and simplicity of the chemical transformations required to generate the compounds of core metabolism imply the possibility of production and assembly of these 24 compounds in the absence of macromolecular catalysts. Some of these 24 compounds such as proline (Pizzarello and Weber, 2004;Limbach, 2005), histidine (Shimizu, 2007; Shimizu et al., 2008), glycine (Plankensteiner et al., 2002), cysteine (Shimizu, 2007; Shimizu et al., 2008), tryptophan (Jiang et al., 2006) also may play the role of small molecule catalysts. Furthermore, it is also possible that other pathway intermediates that possess structural features required for small molecule catalysis—for example the purine pathway intermediates that contain the imidazole moiety—may also participate in such catalytic processes. The simplicity of the chemical transformations and the recursive nature of utilizing a rather small set of rudimentary reactions suggest that the origin, evolution, and organization of the early assembly of the core metabolism might have emerged in an environment conducive to limited diversity in chemical reactions. A primary question of biogenesis is how the anabolic network is generated with great fidelity from the five starting molecular species. A second question is how the relations between amino acids and ribonucleotides emerged (Copley et al., 2005)Conclusion
The complete sequencing of microbial genomes and the availability of various biochemical databases has made it possible to formulate and study the entire metabolic network of many taxa and to compare anabolism across the taxa. This has permitted a detailed study of the metabolism of reductive autotrophs in greater detail than was possible without the sequence data. With this in mind, we reconstructed the complete anabolic chart of intermediary metabolism of a representative reductive autotroph (Srinivasan and Morowitz, 2009). Extending our focus to other kinds of autotrophs, including photo-autotrophs, led us to deconstruct our earlier finding to a canonical network of autotrophic anabolism. This canonical network provides a biochemical universal, which becomes an ecological universal applied to large-scale ecosystems which, overall, are autotrophic in nature. Thus a search motivated by an understanding of biogenesis leads to a broader understanding of all of biology.1) http://www.biolbull.org/content/217/3/222/F1.expansion.html