https://reasonandscience.catsboard.com/t2117-nuclear-pore-complexes-design-or-evolution
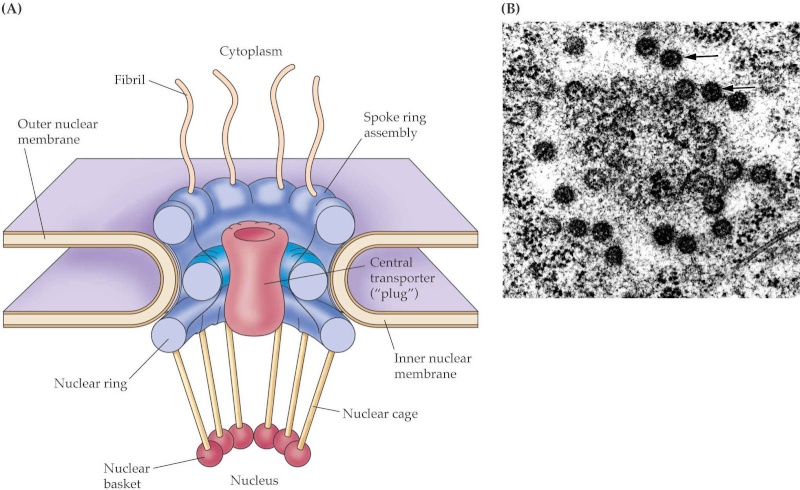
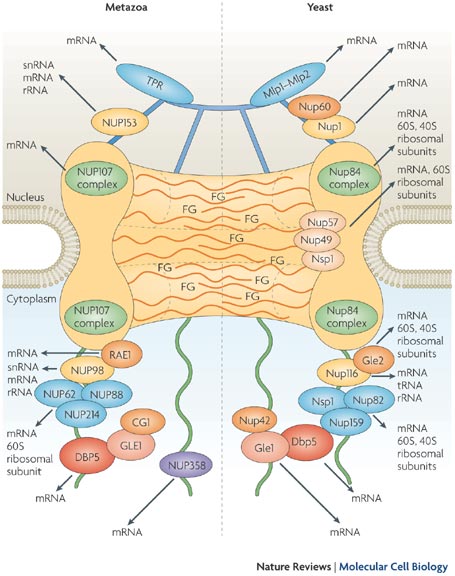
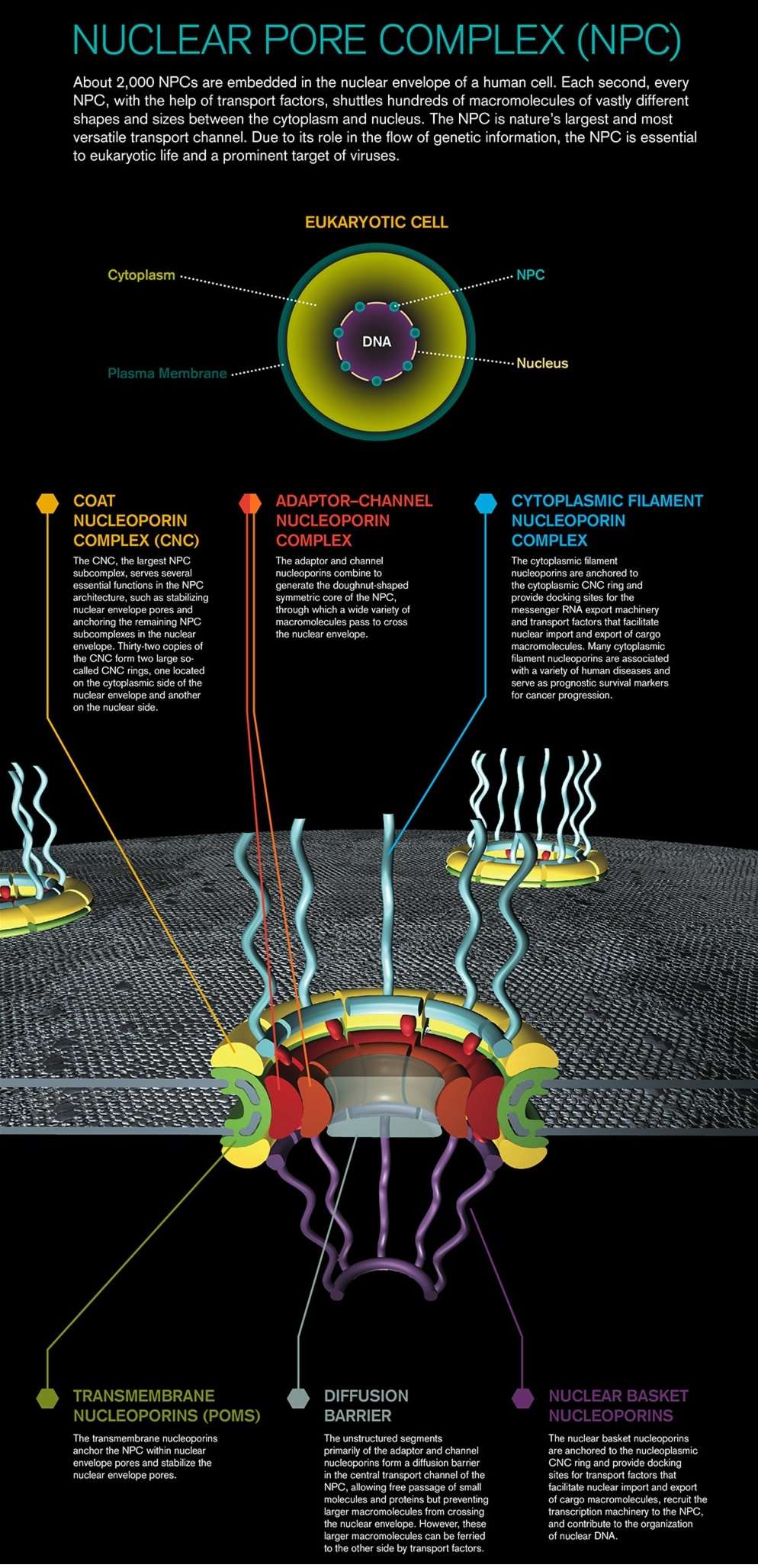
Nuclear pores are large protein complexes that cross the nuclear envelope, which is the double membrane surrounding the eukaryotic cell nucleus. There are about an average of 2000 nuclear pore complexes (NPCs), in the nuclear envelope of a vertebrate cell, but it varies depending on cell type and the stage in the life cycle. The proteins that make up the nuclear pore complex are known as nucleoporins. Each NPC contains at least 456 individual protein molecules and is composed of 30 distinct proteins (nucleoporins). The other half show structural characteristics typical of "natively unfolded" or intrinsically disordered proteins, i.e. they are highly flexible proteins that lack ordered secondary structure.These disordered proteins are the FG nucleoporins, so called because their amino-acid sequence contains many phenylalanine—glycine repeats. 8
Nuclear Pore Complexes Perforate the Nuclear Envelope
Large and elaborate nuclear pore complexes (NPCs) perforate the nuclear envelope in all eukaryotes. Each NPC is composed of a set of approximately 30 different
proteins, or nucleoporins. Reflecting the high degree of internal symmetry, each nucleoporin is present in multiple copies, resulting in 500–1000 protein molecules
in the fully assembled NPC, with an estimated mass of 66 million daltons in yeast and 125 million daltons in vertebrates
Most nucleoporins are composed of repetitive protein domains of only a few different types. Some of the scaffold nucleoporins. The nuclear envelope of a typical mammalian cell contains 3000–4000 NPCs, although that number varies widely, from a few hundred in glial cells to almost 20,000 in Purkinje neurons. The total traffic that passes through each NPC is enormous: each NPC can transport up to 1000 macromolecules per second and can transport in both directions at the same time. How it coordinates the bidirectional flow of macromolecules to avoid congestion and head-on collisions is not known. Each NPC contains aqueous passages, through which small water-soluble molecules can diffuse passively. Researchers have determined the effective size of these passages by injecting labeled water-soluble molecules of different sizes into the cytosol and then measuring their rate of diffusion into the nucleus. Small molecules (5000 daltons or less) diffuse in so fast that we can consider the nuclear envelope freely permeable to them. Large proteins, however, diffuse in much more slowly, and the larger a protein, the more slowly it passes through the NPC. Proteins larger than 60,000 daltons cannot enter by passive diffusion. This size
cut-off to free diffusion is thought to result from the NPC structure. The channel nucleoporins with extensive unstructured regions form a disordered tangle (much like a kelp bed in the ocean) that restricts the diffusion of large macromolecules while allowing smaller molecules to pass. Because many cell proteins are too large to diffuse passively through the NPCs, the nuclear compartment and the cytosol can maintain different protein compositions. Mature cytosolic ribosomes, for example, are about 30 nm in diameter and thus cannot diffuse through the NPC, confining protein synthesis to the cytosol. But how does the nucleus export newly made ribosomal subunits or import large molecules, such as DNA polymerases and RNA polymerases, which have subunit molecular masses of 100,000–200,000 daltons? As we discuss next, these and most other transported protein and RNA molecules bind to specific receptor proteins that actively ferry large molecules through NPCs. Even small proteins like histones frequently use receptor-mediated mechanisms to cross the NPC, thereby increasing transport efficiency.
Nuclear Pore Complexes are elaborate, specialized pores in the nuclear membranes that surround the nucleus of each cell in your body like a skin. The pores look something like complex basketball hoops with rings and studs that act like electronic gates. Their job is to control traffic in and out of the nucleus. Each nuclear pore complex works so fast, it can authenticate somewhere between 520 and 1000 pieces of cargo per second. A typical nucleus has about 2000 to 4000 or more of these gates, which are made up of 30 or more very complex proteins. They all have to be disassembled and reassembled every time a cell divides. 6
Where would you start in trying to work out the structure of a macromolecular machine consisting of 456 proteins? Taking a combined experimental and computational approach is one answer.
Consider a 1,000-piece jigsaw puzzle. There are millions of ways in which the pieces might fit together, yet there is only one solution.
This is just amazing and it exemplifies design in the cell very clearly . Watch the interview of the researchers which figured out the structure of the nuclear pore complex below, and how they describe it as extremely complex, but simple at the same time. Remarkable.
http://www.nature.com/nature/videoarchive/cellarchitecture/
The Structure of the Cell Nucleus “Gatekeeper”
The massive NPC, which allows molecules such as RNAs and proteins to move in and out of the nucleus, is one of the largest macromolecular structures in the cell (containing about 5 million atoms). Its structure has been considered a "holy grail" in structural biology for many decades. The researchers from the Massachusetts Institute of Technology (MIT) found that the NPC scaffold forms an open lattice structure, similar to another membrane coating complex, called COPII. That complex has far fewer components and is involved in a completely different cellular process, called vesicular transport, by which materials move in, out and around the cell enclosed in vesicles.
The finding reveals a remarkable evolutionary story: The proteins that form the building blocks in the NPC and the COPII vesicle coats are so unique that they still have not been identified anywhere else in the cell. Once the models of the proteins were complete, it became clear that these two cellular structures share a common ancestor, dating back over one billion years, which already contained the same building blocks connected in very similar fashion. Second, discovering the specific relationship between the NPC and the much simpler, better understood COPII vesicle coat constitutes a significant step toward understanding the entire NPC assembly, which appears to be remarkably modular and most likely self-assembling.
Details Of Nuclear Pore Complex With Spin 1
A cell’s membrane-bound nucleus uses hundreds to thousands of nuclear pores as its gatekeepers, selective membrane channels that are responsible for regulating the material that goes to and from a cell’s DNA. Rockefeller scientists have nailed down the first complete molecular picture of this huge, 450-protein pore and their findings provide a glimpse into how the nucleus itself first
The group gathered and analyzed massive amounts of data to come up with a rough draft of the structure of the nuclear pore.
The scientists’ results have given them a peek into the early
“We think that once the cells gained this coating complex, they ran with it and started to duplicate it and specialize it,”
“Evolution is a process of duplication and divergence,” Rockefeller professor Michael Rout says. He and his colleagues saw clear evidence of this. For every protein, there was another one that looked quite similar. “These are evidence of duplication events, showing that the evolution of the complicated nuclear pore was a more straightforward affair than previously thought. It’s made of many different variations of a theme of just one unit.”
“The nuclear pore is the communication device that the nucleus uses to communicate with the rest of the cell. And if you don’t understand how that works, you don’t understand a key part of how the cell works. You have to see the cell as a machine and understand all of its parts.”
The authors state:
A cell’s membrane-bound nucleus uses hundreds to thousands of nuclear pores as its gatekeepers, selective membrane channels that are responsible for regulating the material that goes to and from a cell’s DNA.
Molecular Machine Turns Packaged Messenger RNA Into A Linear Transcript 23
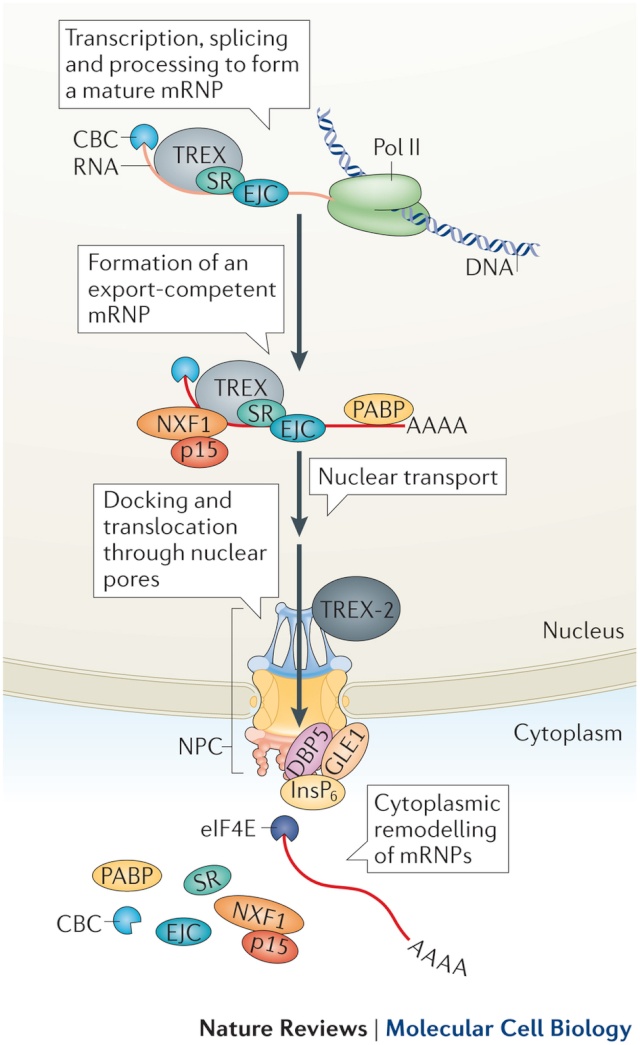
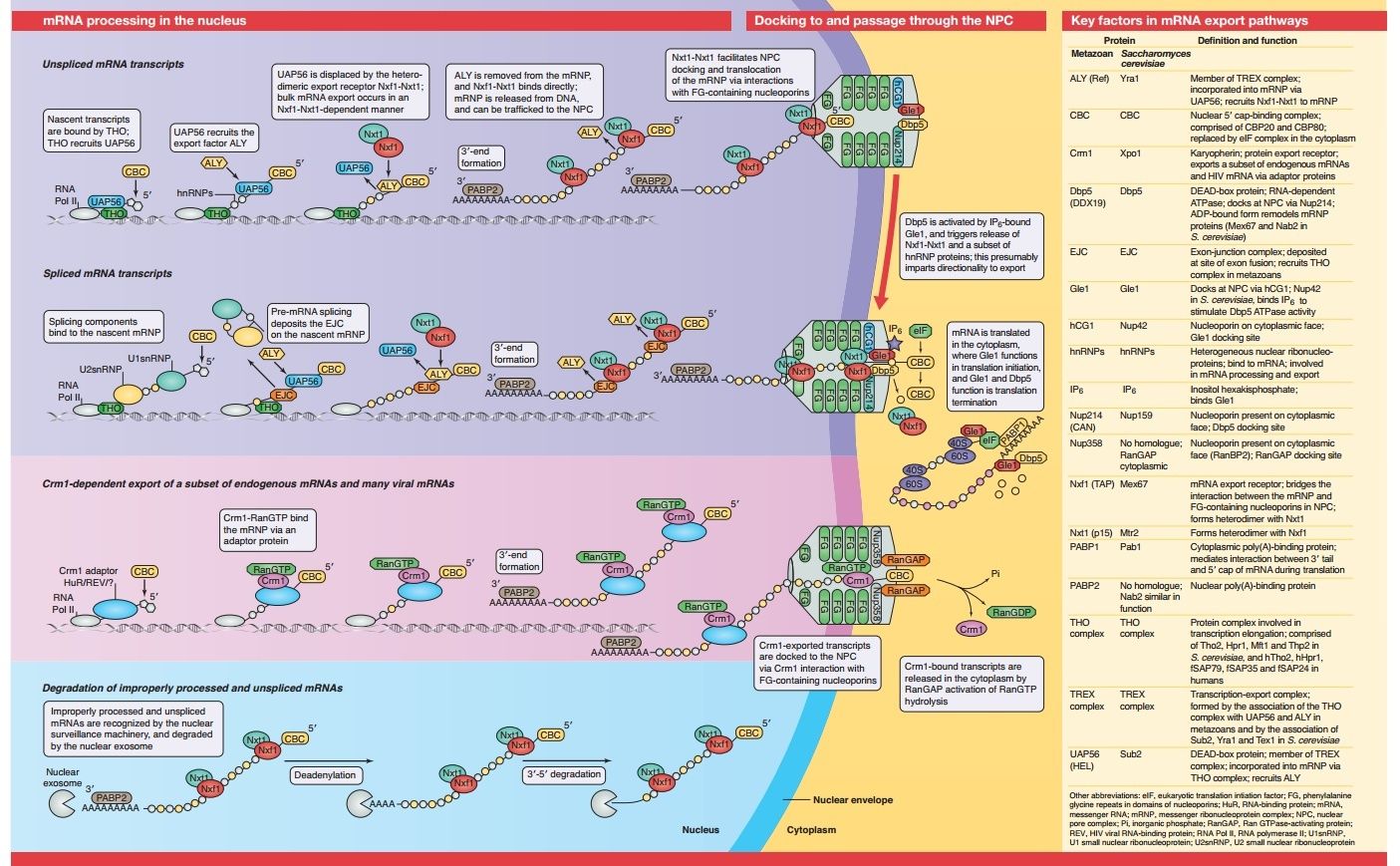
For RNA, the gateway to a productive life outside the nucleus is the nuclear pore complex, an amalgamation of 30 kinds of proteins that regulates all traffic passing through the nuclear membrane. New research shows that one of these proteins magnetically couples with a special molecule -- a helicase -- to form a machine that unpacks balled-up messenger RNA particles so that they can be translated.
The work illuminates a previously unknown stage in the process by which genetic information is read and converted to proteins. In humans and other higher organisms, the genetic information that is encoded in the DNA is stored inside the nucleus, while the factories that convert DNA instructions into proteins are located in the surrounding cytoplasm. As those instructions — messenger RNA particles — pass through the nuclear membrane, numerous proteins that cover and protect the delicate messenger RNA molecules must be stripped off.
André Hoelz, a research associate in John D. Rockefeller Jr. Professor Günter Blobel’s Laboratory of Cell Biology, and his colleagues solved the crystal structure of a complex located on the cytoplasmic side of the nuclear pore — nucleoportin Nup214 coupled with helicase Ddx19. They then performed a series of biochemical experiments to further parse the interactions between these two molecules and to elucidate their mechanism of action. “We found that the messenger RNA protein package and Nup214 competitively bind to the helicase, one after the other,” Hoelz notes. Each time the helicase binds the ball of messenger RNA and protein, it strips one protein molecule off. “The process is akin to a ratchet mechanism for messenger RNA export.” The result, Hoelz speculates, is a linear messenger RNA transcript that travels on to the ribosome, where it delivers instructions for building proteins.
Science daily refers to coordination of independent parts. DNA transcripts made of messenger RNA emerge from the nucleus in 3-D clumps. These need to be “straightened out” into a linear code that can be read by the ribosome. Research at Rockefeller University shows that one of the 30 kinds of proteins in the nuclear pore complex “magnetically” attaches to the transcript when it passes through the gate, joining an unwrapping machine called a helicase “to form a machine that unpacks balled-up messenger RNA particles so that they can be translated.” Here’s how Andre Hoelz described the action: “We found that the messenger RNA protein package and Nup214 competitively bind to the helicase, one after the other.” Each binding strips one protein off as it passes through. “The process is akin to a ratchet mechanism for messenger RNA export,” Hoelz said. Failures in the mechanism, again, were said to be implicated in disease. Once again, also, the article said nothing about evolution. 5
Structural and functional analysis of the interaction between the nucleoporin Nup214 and the DEAD-box helicase Ddx19
Key steps in the export of mRNA from the nucleus to the cytoplasm are the transport through the nuclear pore complex (NPC) and the subsequent remodeling of messenger RNA-protein (mRNP) complexes that occurs at the cytoplasmic side of the NPC. Crucial for these events is the recruitment of the DEAD-box helicase Ddx19 to the cytoplasmic filaments of the NPC that is mediated by the nucleoporin Nup214. Here, we present the crystal structure of the Nup214 N-terminal domain in complex with Ddx19 in its ADP-bound state at 2.5 Å resolution. Strikingly, the interaction surfaces are not only conserved but also exhibit strongly opposing surface potentials, with the helicase surface being positively and the Nup214 surface being negatively charged. We speculate that the positively charged surface of the interacting ADP-helicase binds competitively to a segment of mRNA of a linearized mRNP, passing through the NPC on its way to the cytoplasm. As a result, the ADP-helicase would dissociate from Nup214 and replace a single bound protein from the mRNA. One cycle of protein replacement would be accompanied, cooperatively, by nucleotide exchange, ATP hydrolysis, release of the ADP-helicase from mRNA and its rebinding to Nup214. Repeat of these cycles would remove proteins from a mRNP, one at a time, akin to a ratchet mechanism for mRNA export.
Can evolution explain the origin of the Nuclear Pore Complex? 7
After nine years of work, three universities including a team at Rockefeller University completed a beautiful new model of the nuclear pore complex. The story is told by Science Daily. The article attributed the origin of this exquisite gatekeeper of the nucleus to evolution: “their findings provide a glimpse into how the nucleus itself first evolved,” the article says. How can this be? The article claims that eukaryotic cells split off from the prokaryotes “when they developed a nucleus and other specialized organelles that allowed them to compartmentalize different aspects of cellular metabolism.”
This language suggests some kind of plan or intention – certainly not what neo-Darwinian theory allows. Nevertheless, Michael Rout of Rockefeller continued the language of planning and purpose as he described himself visualizing an evolutionary progress from earlier structures: “We think that once the cells gained this coating complex, they ran with it and started to duplicate it and specialize it.”
As for a mechanism that could generate complexity from simplicity, Rout invoked a kind of copying and tinkering algorithm: “Evolution is a process of duplication and divergence,” he said. Because the nuclear pore complex contains multiple copies of similar structures arranged like a wheel, he thinks this is evidence they must have formed by gene duplication, even though wheels are usually built by intelligent agents:
He and his colleagues saw clear evidence of this when they color-coded the proteins in the pore. One method of color coding revealed alternating stripes, like spokes on a wheel: For every protein, there was another one that looked quite similar. Color coding a different way showed the same pattern in the pore’s outer and inner rings, one of which appears to be a slightly modified duplication of the other. These are evidence of duplication events, Rout says, showing that the evolution of the complicated nuclear pore was a more straightforward affair than previously thought. “It’s made of many different variations of a theme of just one unit.”
mRNA is needed to make the nuclear pore complex. But without the nuclear pore complex, mRNA cannot be prepared for translation in the Ribosome. Thats a catch22 situation....
If this observation is correct, to regulate, there must be:
a sensing mechanism
a feedback mechanism
a control mechanism.
a value against which the parameter is controlled
https://www.youtube.com/watch?v=dJLeLRXIOj0


1) http://www.uncommondescent.com/intelligent-design/details-of-nuclear-pore-complex-with-spin/
2) http://www.sciencedaily.com/releases/2009/02/090213113340.htm
3) http://newswire.rockefeller.edu/2009/02/10/molecular-machine-turns-packaged-messenger-rna-into-a-linear-transcript/
4) http://www.pnas.org/content/106/9/3089.full
5) http://creationsafaris.com/crev200902.htm
6) http://creationsafaris.com/crev0603.htm
7) http://creationsafaris.com/crev200802.htm#20080202a
8 )https://en.wikipedia.org/wiki/Nuclear_pore
further readings :
http://www.nature.com/ncomms/2015/150114/ncomms6978/full/ncomms6978.html
http://www.ks.uiuc.edu/Research/npc/
Last edited by Admin on Mon Feb 04, 2019 4:36 pm; edited 16 times in total