Topoisomerase II enzymes, amazing evidence of design
https://reasonandscience.catsboard.com/t2111-topoisomerase-ii-enzymes-amazing-evidence-of-design
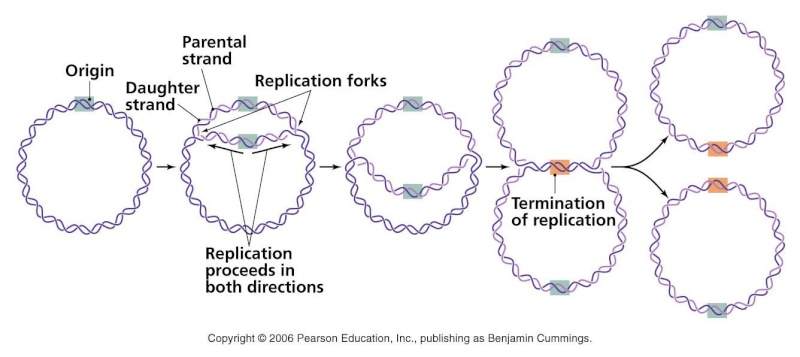
Complete and equal transmission of DNA to daughter cells is crucial during mitosis. During cell division, each daughter cell inherits one copy of every chromosome. The metaphase-to-anaphase transition is the critical point in the cell cycle where the cell commits to separation of sister chromatids. Once spindle attachment is complete, cohesion must be eliminated to enable the physical separation of sister chromatids. This requires cleavage of the protein complex cohesin by separase and, in some instances, completion of chromosome decatenation. Catenation is the process by which two circular DNA strands are linked together like chain links. This occurs after DNA replication, where two single strands are catenated and can still replicate but cannot separate into the two daughter cells.
II Topoisomerase enzymes is a ubiquitous enzyme that is essential for the survival of all eukaryotic organisms and plays critical roles in virtually every aspect of DNA metabolism. It performs the amazing feat of breaking a DNA double helix, passing another helix through the gap, and resealing the double helix behind it. They are essential in the separation of entangled daughter strands during replication. This function is believed to be performed by topoisomerase II in eukaryotes and by topoisomerase IV in prokaryotes. Failure to separate these strands leads to cell death. As genetic material DNA is wonderful, but as a macromolecule, it is unruly, voluminous, and fragile. Without the action of DNA replicases, topoisomerases, helicases, translocases, and recombinases, the genome would collapse into a topologically entangled random coil that would be useless to the cell. The topoisomerase is thought to be a highly dynamic structure, with several gates for entry of DNA into the two DNA-sized holes. Loss of topoisomerase activity in metaphase leads to delayed exit and extensive anaphase chromosome bridging, often resulting in cytokinesis failure, although the maintenance of limited catenation until anaphase may be important for sister chromatid structural organization 9 Accurate transmission of chromosomes requires that the sister DNA molecules created during DNA replication are disentangled and then pulled to opposite poles of the cell before division. Defects in chromosome segregation produce cells that are aneuploid (containing an abnormal number of chromosomes)-a situation that can have dire consequences.
Like many other enzymes, topoisomerase II are essential for cell function, and had to be present in the first living cell to exercise their function right in the beginning, when life began.
Within each chromosome, two dimensions of organization are at play: condensation along the axes ensures the entire chromatid, end-to-end, is kept together 8 , while the tight association of sister chromatids until anaphase, termed sister chromatid cohesion (SCC), ensures that each daughter cell receives only one copy . Two mechanisms are known to play a role in SCC: DNA catenation, which physically interlocks (catenates) DNA across the sister chromatids ; and protein linkages through the cohesin complex, which physically tether the sister chromatids to one another.
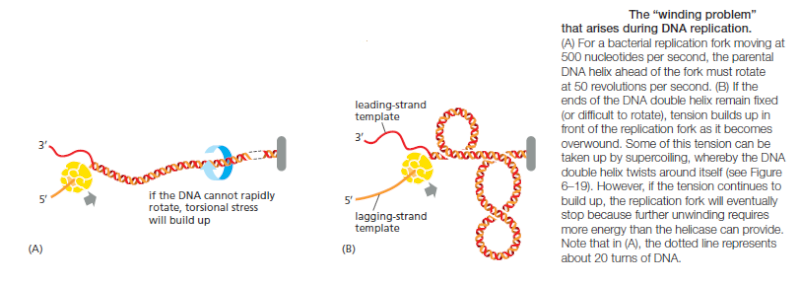
Topoisomerase II forms a covalent linkage to both strands of the DNA helix at the same time, making a transient double-strand break in the helix. These enzymes are activated by sites on chromosomes where two double helices cross over each other such as those generated by supercoiling in front of a replication fork
Once a topoisomerase II molecule binds to such a crossing site, the protein uses ATP hydrolysis to perform the following set of reactions efficiently:
(1) it breaks one double helix reversibly to create a DNA “gate”;
(2) it causes the second, nearby double helix to pass through this opening; and
(3) it then reseals the break and dissociates from the DNA. At crossover points generated by supercoiling, passage of the double helix through the gate occurs in the direction that will reduce supercoiling. In this way, type II topoisomerases can relieve the overwinding tension generated in front of a replication fork. Their reaction mechanism also allows type II DNA topoisomerases to efficiently separate two interlocked DNA circles. Topoisomerase II also prevents the severe DNA tangling problems that would otherwise arise during DNA replication. The enormous usefulness of topoisomerase II for untangling chromosomes can readily be appreciated by anyone who has struggled to remove a tangle from a fishing line without the aid of scissors.
These molecular machines are far beyond what unguided processes involving chance and necessity can produce. Indeed, machinery of the complexity and sophistication of Topoisomerase enzymes are, based on our experience, usually atributed to intelligent agents.
Type IIA topoisomerases consist of several key motifs: an
N-terminal GHKL ATPase domain
Toprim domain
central DNA-binding core
C-terminal domain
Each of these key motifs are essential for the proper function of the enzyme. No part can be reduced, and neither is it possible any of the subparts to emerge by natural means. Not only had the enzyme to emerge prior to the first cell being formed, and so could not be the result of evolution, but the sub parts by themself, and the enzyme by itself even fully formed, would have no use, unless the DNA double helix molecules were already existing as well, and so the whole process of cell division, mitosis, and catenation, which happens through DNA replication. The enzyme is however essential for life, so if Topo II is removed, life could not exist. So we have here one of inumerous essential seemingly tiny and aparently unimportant parts, which by closer looking reveal to be life essential. This provides another big question mark in regard of naturalistic explanations, provides on the other part ones more a powerful argument for design.
http://reasonandscience.heavenforum.org/t2111-topoisomerase-ii-enzymes-amazing-evidence-of-design#3754
Introduction:
The eukaryotic cell is a prime example of a functioning nano machinery. 9
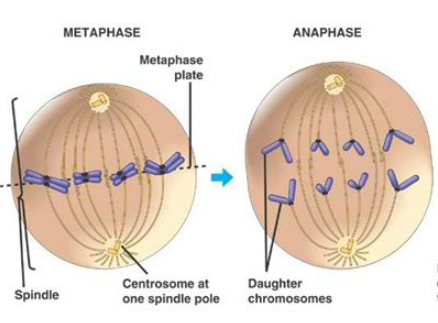
Complete and equal transmission of DNA to daughter cells is crucial during mitosis. During cell division, each daughter cell inherits one copy of every chromosome. The metaphase-to-anaphase transition ( see picture above ) is the critical point in the cell cycle where the cell commits to separation of sister chromatids 2. Once spindle attachment is complete, cohesion must be eliminated to enable the physical separation of sister chromatids. This requires cleavage of the protein complex cohesin by separase and, in some instances, completion of chromosome decatenation. Catenation is the process by which two circular DNA strands are linked together like chain links. This occurs after DNA replication, where two single strands are catenated and can still replicate but cannot separate into the two daughter cells. ( see picture below)
II Topoisomerase enzymes is a ubiquitous enzyme that is essential for the survival of all eukaryotic organisms and plays critical roles in virtually every aspect of DNA metabolism 5 It performs the amazing feat of breaking a DNA double helix, passing another helix through the gap, and resealing the double helix behind it. They are essential in the separation of entangled daughter strands during replication. This function is believed to be performed by topoisomerase II in eukaryotes and by topoisomerase IV in prokaryotes. Failure to separate these strands leads to cell death. As genetic material DNA is wonderful, but as a macromolecule it is unruly, voluminous and fragile. Without the action of DNA replicases, topoisomerases, helicases, translocases and recombinases, the genome would collapse into a topologically entangled random coil that would be useless to the cell. 3 The topoisomerase is thought to be a highly dynamic structure, with several gates for entry of DNA into the two DNA-sized holes. Loss of topoisomerase activity in metaphase leads to delayed exit and extensive anaphase chromosome bridging, often resulting in cytokinesis failure, although maintenance of limited catenation until anaphase may be important for sister chromatid structural organization 9 Accurate transmission of chromosomes requires that the sister DNA molecules created during DNA replication are disentangled and then pulled to opposite poles of the cell before division. Defects in chromosome segregation produce cells that are aneuploid (containing an abnormal number of chromosomes)-a situation that can have dire consequences. 7
Like many other enzymes, topoisomerase II are essential for cell function, and had to be present in the first living cell to exercise their function right in the beginning, when life began.
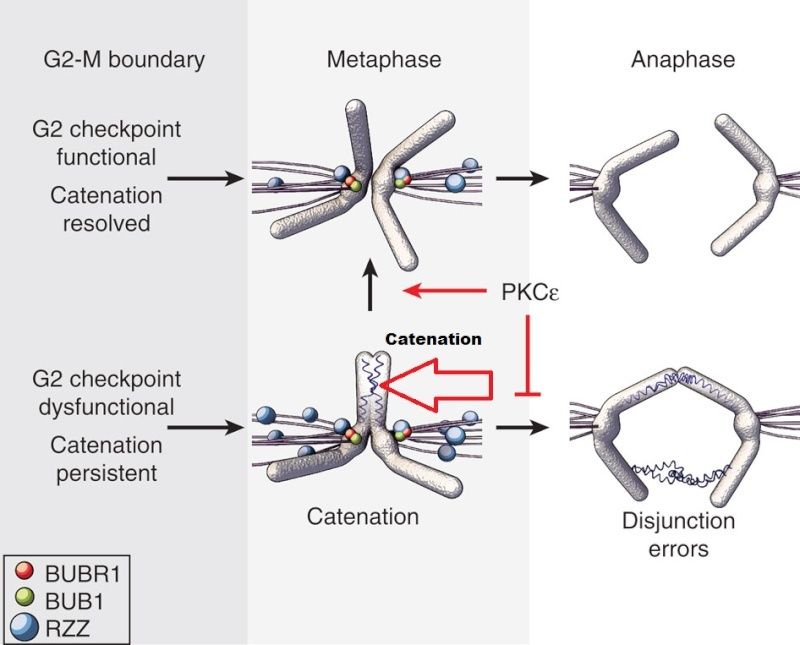
Within each chromosome, two dimensions of organization are at play: condensation along the axes ensures the entire chromatid, end-to-end, is kept together 8 , while the tight association of sister chromatids until anaphase, termed sister chromatid cohesion (SCC), ensures that each daughter cell receives only one copy . Two mechanisms are known to play a role in SCC: DNA catenation, which physically interlocks (catenates) DNA across the sister chromatids ; and protein linkages through the cohesin complex, which physically tether the sister chromatids to one another.
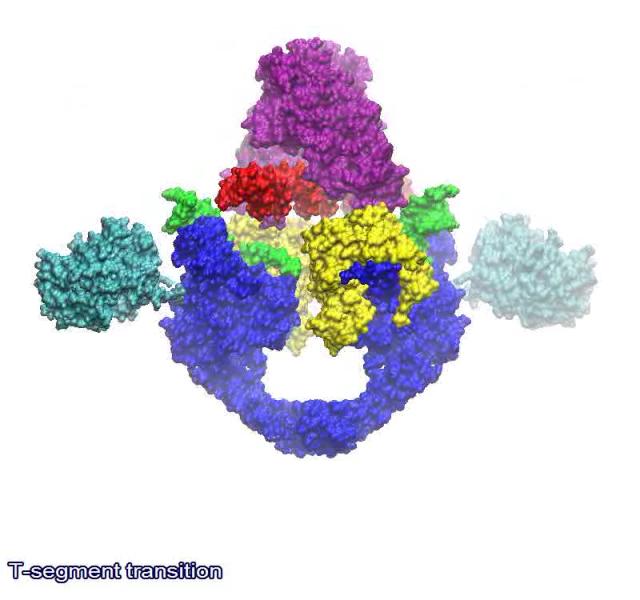
The link below shows a animated model of the DNA transport mechanism employed by a type II DNA topoisomerases. N-terminal domain of ParC is shown in blue, C-terminal domain of ParC in cyan, N-terminal domain of ParE in purple, C-terminal domain of ParE in yellow, G-segment in green and the DNA strand in red.
https://upload.wikimedia.org/wikipedia/commons/8/8b/Structural-Basis-of-Gate-DNA-Breakage-and-Resealing-by-Type-II-Topoisomerases-pone.0011338.s005.ogv
https://bioslawek.files.wordpress.com/2011/09/numern-5.gif?w=450&h=450
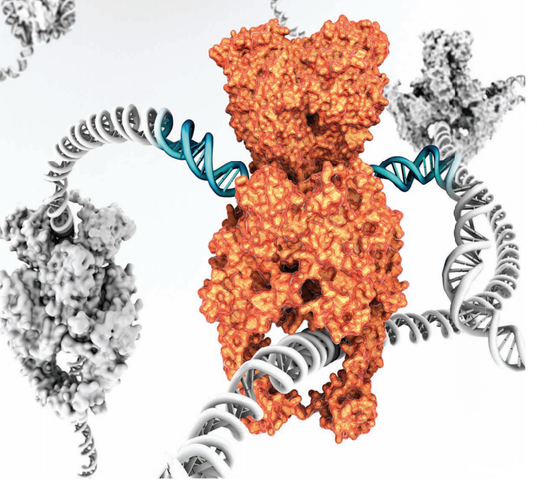
Type II topoisomerases cut both strands of the DNA helix simultaneously in order to manage DNA tangles and supercoils. They use the hydrolysis of ATP 1
Sister-chromatid cohesion also results, at least in part, from DNA catenation, the intertwining of sister DNA molecules that occurs when two replication forks meet during DNA synthesis.
The enzyme topoisomerase II gradually disentangles the catenated sister DNAs between
S phase and early mitosis by cutting one DNA molecule, passing the other through the break,
and then resealing the cut DNA
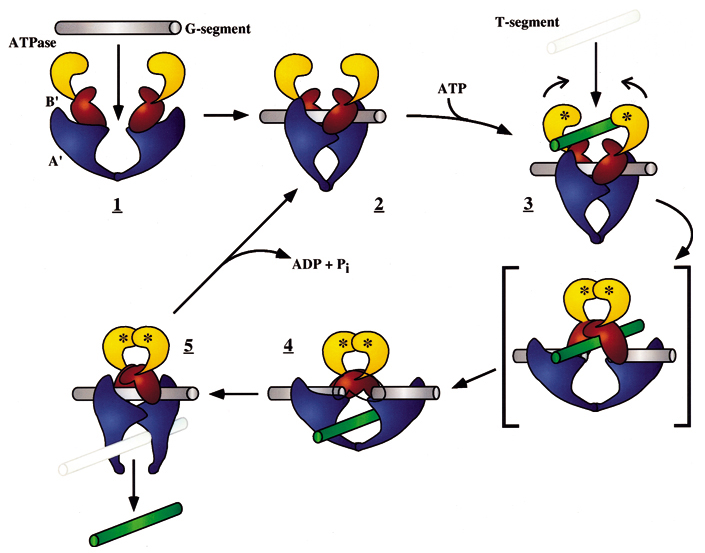
Picture below: The DNA-helix-passing reaction catalyzed by DNA topoisomerase II. type II enzymes hydrolyze ATP (red), which is needed to release and reset the enzyme after each cycle. Type II topoisomerases are largely confined to proliferating cells in eukaryotes; partly for that reason, they have been effective targets for anticancer drugs. Some of these drugs inhibit topoisomerase II at the third step in the figure and thereby produce high levels of double-strand breaks that kill rapidly dividing cells. The small yellow circles represent the phosphates in the DNA backbone that become covalently bonded to the topoisomerase
Topoisomerase II forms a covalent linkage to both strands of the DNA helix at the same time, making a transient double-strand break in the helix. These enzymes are activated by sites on chromosomes where two double helices cross over each other such as those generated by supercoiling in front of a replication fork
Once a topoisomerase II molecule binds to such a crossing site, the protein uses ATP hydrolysis to perform the following set of reactions efficiently:
(1) it breaks one double helix reversibly to create a DNA “gate”;
(2) it causes the second, nearby double helix to pass through this opening; and
(3) it then reseals the break and dissociates from the DNA. At crossover points generated by supercoiling, passage of the double helix through the gate occurs in the direction that will reduce supercoiling. In this way, type II topoisomerases can relieve the overwinding tension generated in front of a replication fork. Their reaction mechanism also allows type II DNA topoisomerases to efficiently separate two interlocked DNA circles. Topoisomerase II also prevents the severe DNA tangling problems that would otherwise arise during DNA replication. The enormous usefulness of topoisomerase II for untangling chromosomes can readily be appreciated by anyone who has struggled to remove a tangle from a fishing line without the aid of scissors.
These molecular machines are far beyond what unguided processes involving chance and necessity can produce. Indeed, machinery of the complexity and sophistication of Topoisomerase enzymes are, based on our experience, usually attributed to intelligent agents.
"Creationists Let Facts Speak for Themselves in Mainstream Science Publication"
Although this publication uses an expression I detest (facts do not speak for themselves, they are interpreted), it is worth noting that in a rare occasion, a creationists was allowed to participate in scientific research.
*****
Joseph Deweese has so far managed to continue as a mainstream scientist while openly professing his belief in creation. He is simultaneously an associate professor of Biochemistry at Lipscomb University and an adjunct professor at Vanderbilt University School of Medicine.
Deweese has published in various top science journals (including Nature) on the Topoisomerase family of enzymes. He (with yours truly) recently published a paper in the journal FASEB (Federation of American Societies for Experimental Biology) in connection with the conference on Experimental Biology in Orlando, Florida, this past April.
https://crev.info/2019/06/creationist-topoisomerase-research/
https://www.youtube.com/watch?v=JSM6U32AVyc
1) https://en.wikipedia.org/wiki/Type_II_topoisomerase
2) http://www.rcsb.org/pdb/101/motm.do?momID=73
3) http://www.nature.com/nrm/journal/v7/n8/full/nrm1982.html
4) http://www.ncbi.nlm.nih.gov/books/NBK21703/
5) http://www.sciencedirect.com/science/article/pii/S0167478198001328
6) https://en.wikipedia.org/wiki/DNA_supercoil
7) http://www.ncbi.nlm.nih.gov/pubmed/12142526
8 )http://reasonandscience.heavenforum.org/t2086-chromosome-condensation-amazing-evidence-of-design
9) http://foresight.org/Conference/MNT6/Abstracts/Knoch/index.html
10) http://www.ncbi.nlm.nih.gov/pubmed/17293019
https://reasonandscience.catsboard.com/t2111-topoisomerase-ii-enzymes-amazing-evidence-of-design
Complete and equal transmission of DNA to daughter cells is crucial during mitosis. During cell division, each daughter cell inherits one copy of every chromosome. The metaphase-to-anaphase transition is the critical point in the cell cycle where the cell commits to separation of sister chromatids. Once spindle attachment is complete, cohesion must be eliminated to enable the physical separation of sister chromatids. This requires cleavage of the protein complex cohesin by separase and, in some instances, completion of chromosome decatenation. Catenation is the process by which two circular DNA strands are linked together like chain links. This occurs after DNA replication, where two single strands are catenated and can still replicate but cannot separate into the two daughter cells.
II Topoisomerase enzymes is a ubiquitous enzyme that is essential for the survival of all eukaryotic organisms and plays critical roles in virtually every aspect of DNA metabolism. It performs the amazing feat of breaking a DNA double helix, passing another helix through the gap, and resealing the double helix behind it. They are essential in the separation of entangled daughter strands during replication. This function is believed to be performed by topoisomerase II in eukaryotes and by topoisomerase IV in prokaryotes. Failure to separate these strands leads to cell death. As genetic material DNA is wonderful, but as a macromolecule, it is unruly, voluminous, and fragile. Without the action of DNA replicases, topoisomerases, helicases, translocases, and recombinases, the genome would collapse into a topologically entangled random coil that would be useless to the cell. The topoisomerase is thought to be a highly dynamic structure, with several gates for entry of DNA into the two DNA-sized holes. Loss of topoisomerase activity in metaphase leads to delayed exit and extensive anaphase chromosome bridging, often resulting in cytokinesis failure, although the maintenance of limited catenation until anaphase may be important for sister chromatid structural organization 9 Accurate transmission of chromosomes requires that the sister DNA molecules created during DNA replication are disentangled and then pulled to opposite poles of the cell before division. Defects in chromosome segregation produce cells that are aneuploid (containing an abnormal number of chromosomes)-a situation that can have dire consequences.
Like many other enzymes, topoisomerase II are essential for cell function, and had to be present in the first living cell to exercise their function right in the beginning, when life began.
Within each chromosome, two dimensions of organization are at play: condensation along the axes ensures the entire chromatid, end-to-end, is kept together 8 , while the tight association of sister chromatids until anaphase, termed sister chromatid cohesion (SCC), ensures that each daughter cell receives only one copy . Two mechanisms are known to play a role in SCC: DNA catenation, which physically interlocks (catenates) DNA across the sister chromatids ; and protein linkages through the cohesin complex, which physically tether the sister chromatids to one another.
Topoisomerase II forms a covalent linkage to both strands of the DNA helix at the same time, making a transient double-strand break in the helix. These enzymes are activated by sites on chromosomes where two double helices cross over each other such as those generated by supercoiling in front of a replication fork
Once a topoisomerase II molecule binds to such a crossing site, the protein uses ATP hydrolysis to perform the following set of reactions efficiently:
(1) it breaks one double helix reversibly to create a DNA “gate”;
(2) it causes the second, nearby double helix to pass through this opening; and
(3) it then reseals the break and dissociates from the DNA. At crossover points generated by supercoiling, passage of the double helix through the gate occurs in the direction that will reduce supercoiling. In this way, type II topoisomerases can relieve the overwinding tension generated in front of a replication fork. Their reaction mechanism also allows type II DNA topoisomerases to efficiently separate two interlocked DNA circles. Topoisomerase II also prevents the severe DNA tangling problems that would otherwise arise during DNA replication. The enormous usefulness of topoisomerase II for untangling chromosomes can readily be appreciated by anyone who has struggled to remove a tangle from a fishing line without the aid of scissors.
These molecular machines are far beyond what unguided processes involving chance and necessity can produce. Indeed, machinery of the complexity and sophistication of Topoisomerase enzymes are, based on our experience, usually atributed to intelligent agents.
Type IIA topoisomerases consist of several key motifs: an
N-terminal GHKL ATPase domain
Toprim domain
central DNA-binding core
C-terminal domain
Each of these key motifs are essential for the proper function of the enzyme. No part can be reduced, and neither is it possible any of the subparts to emerge by natural means. Not only had the enzyme to emerge prior to the first cell being formed, and so could not be the result of evolution, but the sub parts by themself, and the enzyme by itself even fully formed, would have no use, unless the DNA double helix molecules were already existing as well, and so the whole process of cell division, mitosis, and catenation, which happens through DNA replication. The enzyme is however essential for life, so if Topo II is removed, life could not exist. So we have here one of inumerous essential seemingly tiny and aparently unimportant parts, which by closer looking reveal to be life essential. This provides another big question mark in regard of naturalistic explanations, provides on the other part ones more a powerful argument for design.
http://reasonandscience.heavenforum.org/t2111-topoisomerase-ii-enzymes-amazing-evidence-of-design#3754
Introduction:
The eukaryotic cell is a prime example of a functioning nano machinery. 9

Complete and equal transmission of DNA to daughter cells is crucial during mitosis. During cell division, each daughter cell inherits one copy of every chromosome. The metaphase-to-anaphase transition ( see picture above ) is the critical point in the cell cycle where the cell commits to separation of sister chromatids 2. Once spindle attachment is complete, cohesion must be eliminated to enable the physical separation of sister chromatids. This requires cleavage of the protein complex cohesin by separase and, in some instances, completion of chromosome decatenation. Catenation is the process by which two circular DNA strands are linked together like chain links. This occurs after DNA replication, where two single strands are catenated and can still replicate but cannot separate into the two daughter cells. ( see picture below)
II Topoisomerase enzymes is a ubiquitous enzyme that is essential for the survival of all eukaryotic organisms and plays critical roles in virtually every aspect of DNA metabolism 5 It performs the amazing feat of breaking a DNA double helix, passing another helix through the gap, and resealing the double helix behind it. They are essential in the separation of entangled daughter strands during replication. This function is believed to be performed by topoisomerase II in eukaryotes and by topoisomerase IV in prokaryotes. Failure to separate these strands leads to cell death. As genetic material DNA is wonderful, but as a macromolecule it is unruly, voluminous and fragile. Without the action of DNA replicases, topoisomerases, helicases, translocases and recombinases, the genome would collapse into a topologically entangled random coil that would be useless to the cell. 3 The topoisomerase is thought to be a highly dynamic structure, with several gates for entry of DNA into the two DNA-sized holes. Loss of topoisomerase activity in metaphase leads to delayed exit and extensive anaphase chromosome bridging, often resulting in cytokinesis failure, although maintenance of limited catenation until anaphase may be important for sister chromatid structural organization 9 Accurate transmission of chromosomes requires that the sister DNA molecules created during DNA replication are disentangled and then pulled to opposite poles of the cell before division. Defects in chromosome segregation produce cells that are aneuploid (containing an abnormal number of chromosomes)-a situation that can have dire consequences. 7
Like many other enzymes, topoisomerase II are essential for cell function, and had to be present in the first living cell to exercise their function right in the beginning, when life began.

Within each chromosome, two dimensions of organization are at play: condensation along the axes ensures the entire chromatid, end-to-end, is kept together 8 , while the tight association of sister chromatids until anaphase, termed sister chromatid cohesion (SCC), ensures that each daughter cell receives only one copy . Two mechanisms are known to play a role in SCC: DNA catenation, which physically interlocks (catenates) DNA across the sister chromatids ; and protein linkages through the cohesin complex, which physically tether the sister chromatids to one another.
The link below shows a animated model of the DNA transport mechanism employed by a type II DNA topoisomerases. N-terminal domain of ParC is shown in blue, C-terminal domain of ParC in cyan, N-terminal domain of ParE in purple, C-terminal domain of ParE in yellow, G-segment in green and the DNA strand in red.
https://upload.wikimedia.org/wikipedia/commons/8/8b/Structural-Basis-of-Gate-DNA-Breakage-and-Resealing-by-Type-II-Topoisomerases-pone.0011338.s005.ogv

https://bioslawek.files.wordpress.com/2011/09/numern-5.gif?w=450&h=450

Type II topoisomerases cut both strands of the DNA helix simultaneously in order to manage DNA tangles and supercoils. They use the hydrolysis of ATP 1
Sister-chromatid cohesion also results, at least in part, from DNA catenation, the intertwining of sister DNA molecules that occurs when two replication forks meet during DNA synthesis.

The enzyme topoisomerase II gradually disentangles the catenated sister DNAs between
S phase and early mitosis by cutting one DNA molecule, passing the other through the break,
and then resealing the cut DNA
Picture below: The DNA-helix-passing reaction catalyzed by DNA topoisomerase II. type II enzymes hydrolyze ATP (red), which is needed to release and reset the enzyme after each cycle. Type II topoisomerases are largely confined to proliferating cells in eukaryotes; partly for that reason, they have been effective targets for anticancer drugs. Some of these drugs inhibit topoisomerase II at the third step in the figure and thereby produce high levels of double-strand breaks that kill rapidly dividing cells. The small yellow circles represent the phosphates in the DNA backbone that become covalently bonded to the topoisomerase

Topoisomerase II forms a covalent linkage to both strands of the DNA helix at the same time, making a transient double-strand break in the helix. These enzymes are activated by sites on chromosomes where two double helices cross over each other such as those generated by supercoiling in front of a replication fork
Once a topoisomerase II molecule binds to such a crossing site, the protein uses ATP hydrolysis to perform the following set of reactions efficiently:
(1) it breaks one double helix reversibly to create a DNA “gate”;
(2) it causes the second, nearby double helix to pass through this opening; and
(3) it then reseals the break and dissociates from the DNA. At crossover points generated by supercoiling, passage of the double helix through the gate occurs in the direction that will reduce supercoiling. In this way, type II topoisomerases can relieve the overwinding tension generated in front of a replication fork. Their reaction mechanism also allows type II DNA topoisomerases to efficiently separate two interlocked DNA circles. Topoisomerase II also prevents the severe DNA tangling problems that would otherwise arise during DNA replication. The enormous usefulness of topoisomerase II for untangling chromosomes can readily be appreciated by anyone who has struggled to remove a tangle from a fishing line without the aid of scissors.
These molecular machines are far beyond what unguided processes involving chance and necessity can produce. Indeed, machinery of the complexity and sophistication of Topoisomerase enzymes are, based on our experience, usually attributed to intelligent agents.
"Creationists Let Facts Speak for Themselves in Mainstream Science Publication"
Although this publication uses an expression I detest (facts do not speak for themselves, they are interpreted), it is worth noting that in a rare occasion, a creationists was allowed to participate in scientific research.
*****
Joseph Deweese has so far managed to continue as a mainstream scientist while openly professing his belief in creation. He is simultaneously an associate professor of Biochemistry at Lipscomb University and an adjunct professor at Vanderbilt University School of Medicine.
Deweese has published in various top science journals (including Nature) on the Topoisomerase family of enzymes. He (with yours truly) recently published a paper in the journal FASEB (Federation of American Societies for Experimental Biology) in connection with the conference on Experimental Biology in Orlando, Florida, this past April.
https://crev.info/2019/06/creationist-topoisomerase-research/
https://www.youtube.com/watch?v=JSM6U32AVyc
1) https://en.wikipedia.org/wiki/Type_II_topoisomerase
2) http://www.rcsb.org/pdb/101/motm.do?momID=73
3) http://www.nature.com/nrm/journal/v7/n8/full/nrm1982.html
4) http://www.ncbi.nlm.nih.gov/books/NBK21703/
5) http://www.sciencedirect.com/science/article/pii/S0167478198001328
6) https://en.wikipedia.org/wiki/DNA_supercoil
7) http://www.ncbi.nlm.nih.gov/pubmed/12142526
8 )http://reasonandscience.heavenforum.org/t2086-chromosome-condensation-amazing-evidence-of-design
9) http://foresight.org/Conference/MNT6/Abstracts/Knoch/index.html
10) http://www.ncbi.nlm.nih.gov/pubmed/17293019
Last edited by Otangelo on Mon Aug 01, 2022 6:00 am; edited 25 times in total