The Cell cycle
https://reasonandscience.catsboard.com/t2109-the-cell-cycle

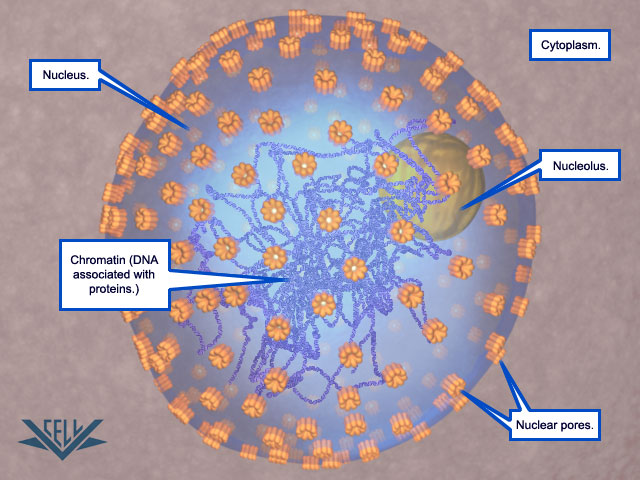
The duplication of eukaryotic cells is a all fine-tuned biochemical processes that depend on the precise structural arrangement of the cellular components. 2
The only way to make a new cell is to duplicate a cell that already exists.
That raises the question how the first cell could have emerged
This simple fact, first established in the middle of the nineteenth century, carries with it a profound message for the continuity of life. All living organisms, from the unicellular bacterium to the multicellular mammal, are products of repeated rounds of cell growth and division extending back in time to the beginnings of life. A cell reproduces by performing an orderly sequence of events in which it duplicates its contents and then divides in two.
Question : how was the right orderly sequence of events achieved ? Is not forsight and order right from the beginning required ?
This cycle of duplication and division, known as the cell cycle, is the essential mechanism by which all living things reproduce. In unicellular species, such as bacteria and yeasts, each cell division produces a complete new organism. In multicellular species, long and complex sequences of cell divisions are required to produce a functioning organism. Even in the adult body, cell division is usually needed to replace cells that die. In fact, each of us must manufacture many millions of cells every second simply to survive: if all cell division were stopped—by exposure to a very large dose of x-rays, for example—we would die within a few days. The details of the cell cycle vary from organism to organism and at different times in an organism’s life. Certain characteristics, however, are universal. At a minimum, the cell must accomplish its most fundamental task: the passing on of its genetic information to the next generation of cells. To produce two genetically identical daughter cells, the DNA in each chromosome must first be faithfully replicated to produce two complete copies. The replicated chromosomes must then be accurately distributed (segregated) to the two daughter cells, so that each receives a copy of the entire genome.
The most basic function of the cell cycle is to duplicate the vast amount of DNA in the chromosomes and then segregate the copies into two genetically identical daughter cells. These processes define the two major phases of the cell cycle. Chromosome duplication occurs during S phase (S for DNA synthesis), which requires 10–12 hours and occupies about half of the cell-cycle time in a typical mammalian cell. After S phase, chromosome segregation and cell division occur in M phase (M for mitosis), which requires much less time (less than an hour in a mammalian cell). M phase comprises two major events: nuclear division, or mitosis, during which the copied chromosomes are distributed into a pair of daughter nuclei; and cytoplasmic division, or cytokinesis, when the cell itself divides in two
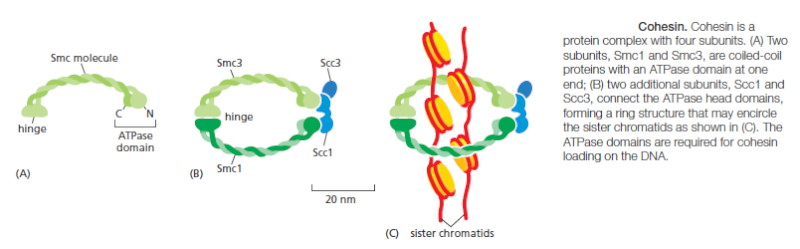
At the end of S phase, the DNA molecules in each pair of duplicated chromosomes are intertwined and held tightly together by specialized protein linkages. Early in mitosis at a stage called prophase, the two DNA molecules are gradually disentangled and condensed into pairs of rigid, compact rods called sister chromatids, which remain linked by sister-chromatid cohesion. When the nuclear envelope disassembles later in mitosis, the sister-chromatid pairs become attached to the mitotic spindle, a giant bipolar array of microtubules . Sister chromatids are attached to opposite poles of the spindle and, eventually, align at the spindle equator in a stage called metaphase. The destruction of sister-chromatid cohesion at the start of anaphase separates the sister chromatids, which are pulled to opposite poles of the spindle. The spindle is then disassembled, and the segregated chromosomes are packaged into separate nuclei at telophase. Cytokinesis then cleaves the cell in two, so that each daughter cell inherits one of the two nuclei.
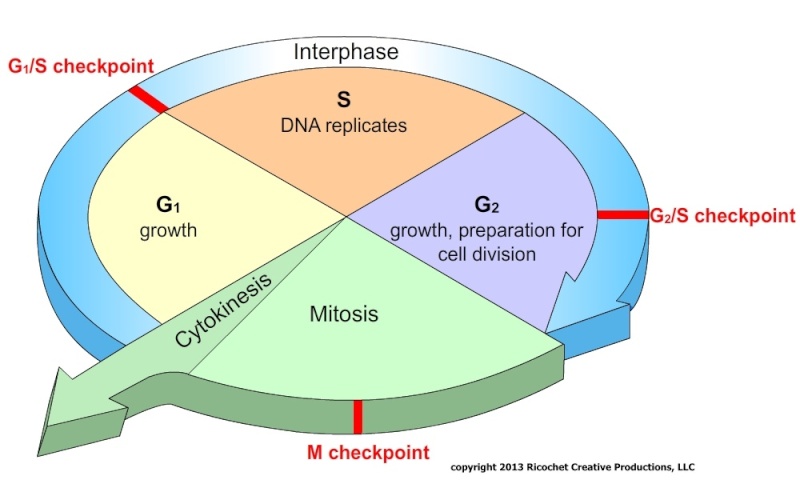


The Eukaryotic Cell Cycle Usually Consists of Four Phases
Most cells require much more time to grow and double their mass of proteins and organelles than they require to duplicate their chromosomes and divide. Partly to allow time for growth, most cell cycles have gap phases—a G1 phase between M phase and S phase and a G2 phase between S phase and mitosis. Thus, the eukaryotic cell cycle is traditionally divided into four sequential phases: G1, S, G2, and M. G1, S, and G2 together are called interphase . In a typical human cell proliferating in culture, interphase might occupy 23 hours of a 24-hour cycle, with 1 hour for M phase. Cell growth occurs throughout the cell cycle, except during mitosis. The two gap phases are more than simple time delays to allow cell growth. They also provide time for the cell to monitor the internal and external environment. to ensure that conditions are suitable and preparations are complete before the cell commits itself to the major upheavals of S phase and mitosis. The G1 phase is especially important in this respect. Its length can vary greatly depending on external conditions and extracellular signals from other cells. If extracellular conditions are unfavorable, for example, cells delay progress through G1 and may even enter a specialized resting state known as G0 (G zero), in which they can remain for days, weeks, or even years before resuming proliferation. Indeed, many cells remain permanently in G0 until they or the organism dies. If extracellular conditions are favorable and signals to grow and divide are present, cells in early G1 or G0 progress through a commitment point near the end of G1 known as Start (in yeasts) or the restriction point (in mammalian cells). We will use the term Start for both yeast and animal cells. After passing this point, cells are committed to DNA replication, even if the extracellular signals that stimulate cell growth and division are removed.
Cell-Cycle Control Is Similar in All Eukaryotes
Some features of the cell cycle, including the time required to complete certain events, vary greatly from one cell type to another, even in the same organism. The basic organization of the cycle, however, is essentially the same in all eukaryotic cells, and all eukaryotes appear to use similar machinery and control mechanisms to drive and regulate cell-cycle events. The proteins of the cell-cycle control system have been so well conserved that many of them function perfectly when transferred from a human cell to a yeast cell. We can therefore study the cell cycle and its regulation in a variety of organisms and use the findings from all of them to assemble a unified picture of how eukaryotic cells divide. Several model organisms are used in the analysis of the eukaryotic cell cycle. The budding yeast Saccharomyces cerevisiae and the fission yeast Schizosaccharomyces pombe are simple eukaryotes in which powerful molecular and genetic approaches can be used to identify and characterize the genes and proteins that govern the fundamental features of cell division. The early embryos of certain animals, particularly those of the frog Xenopus laevis, are excellent tools for biochemical dissection of cell-cycle control mechanisms, while the fruit fly Drosophila melanogaster is useful for the genetic analysis of mechanisms underlying the control and coordination of cell growth and division in multicellular organisms. Cultured human cells provide an excellent system for the molecular and microscopic exploration of the complex processes by which our own cells divide.
Summary
Cell division usually begins with duplication of the cell’s contents, followed by distribution of those contents into two daughter cells. Chromosome duplication occurs during S phase of the cell cycle, whereas most other cell components are duplicated continuously throughout the cycle. During M phase, the replicated chromosomes are segregated into individual nuclei (mitosis), and the cell then splits in two (cytokinesis). S phase and M phase are usually separated by gap phases called G1 and G2, when various intracellular and extracellular signals regulate cell-cycle progression. Cell-cycle organization and control have been highly conserved during evolution, and studies in a wide range of systems have led to a unified view of eukaryotic cell-cycle control
1) http://tfscientist.hubpages.com/hub/Stages-of-the-Cell-Cycle-Mitosis-Part-2-of-2
2) http://foresight.org/Conference/MNT6/Abstracts/Knoch/index.html
https://reasonandscience.catsboard.com/t2109-the-cell-cycle

The duplication of eukaryotic cells is a all fine-tuned biochemical processes that depend on the precise structural arrangement of the cellular components. 2
The only way to make a new cell is to duplicate a cell that already exists.
That raises the question how the first cell could have emerged
This simple fact, first established in the middle of the nineteenth century, carries with it a profound message for the continuity of life. All living organisms, from the unicellular bacterium to the multicellular mammal, are products of repeated rounds of cell growth and division extending back in time to the beginnings of life. A cell reproduces by performing an orderly sequence of events in which it duplicates its contents and then divides in two.
Question : how was the right orderly sequence of events achieved ? Is not forsight and order right from the beginning required ?
This cycle of duplication and division, known as the cell cycle, is the essential mechanism by which all living things reproduce. In unicellular species, such as bacteria and yeasts, each cell division produces a complete new organism. In multicellular species, long and complex sequences of cell divisions are required to produce a functioning organism. Even in the adult body, cell division is usually needed to replace cells that die. In fact, each of us must manufacture many millions of cells every second simply to survive: if all cell division were stopped—by exposure to a very large dose of x-rays, for example—we would die within a few days. The details of the cell cycle vary from organism to organism and at different times in an organism’s life. Certain characteristics, however, are universal. At a minimum, the cell must accomplish its most fundamental task: the passing on of its genetic information to the next generation of cells. To produce two genetically identical daughter cells, the DNA in each chromosome must first be faithfully replicated to produce two complete copies. The replicated chromosomes must then be accurately distributed (segregated) to the two daughter cells, so that each receives a copy of the entire genome.
The Mitotic Cell Cycle
Metaphase, Anaphase and Telophase
Part one of this series looked at the cycles within cycles that make up the existence of a cell. Whilst taking up such a small percentage of the overall cell cycle, mitosis is one of the most important series of events in the life of a cell. 1
Mitosis divides the nucleus of a cell into two new nuclei.
The first stage of mitosis is prophase. Here we see the DNA has wrapped tightly around
proteins to form chromosomes, the nucleolus disappears, and microtubules begin to grow
out from the centrosomes.
The next stage is prometaphase where the nuclear membrane breaks down.
In prometaphase, the microtubules also lengthen by the addition of tubulin proteins to the growing end.
The microtubules then attach to the chromosomes at the kinetochore--a protein complex located at the
centromere of each chromosome.
In prometaphase, the microtubules also lengthen by the addition of tubulin proteins to the
growing end. The microtubules then attach to the chromosomes at the kinetochore--a protein
complex located at the centromere of each chromosome.
In metaphase, the chromosomes align at the equator of the cell.
In anaphase, the sister chromatids are separated. The separation occurs when the microtubules
connected to the chromatids shorten by the loss of tubulin.
Also, the microtubules not connected to the chromatids lengthen to push the two poles of the cell apart.
In telophase, the spindle apparatus made up of microtubules breaks down.
After the spindle apparatus breaks down, the nuclear membrane reforms, and the chromosome uncoil.
Finally, at the end of telophase, the nucleolus reappears. Mitosis is now complete.
In addition to duplicating their genome, most cells also duplicate their other organelles and macromolecules; otherwise, daughter cells would get smaller with each division. To maintain their size, dividing cells must coordinate their growth (that is, their increase in cell mass) with their division. We begin with a brief overview of the cell cycle. We then describe the cell-cycle control system, a complex network of regulatory proteins that triggers the different events of the cycle. We next consider in detail the major stages of the cell cycle, in which the chromosomes are duplicated and then segregated into the two daughter cells. Finally, we consider how extracellular signals govern the rates of cell growth and division and how these two processes are coordinated.
OVERVIEW OF THE CELL CYCLEThe most basic function of the cell cycle is to duplicate the vast amount of DNA in the chromosomes and then segregate the copies into two genetically identical daughter cells. These processes define the two major phases of the cell cycle. Chromosome duplication occurs during S phase (S for DNA synthesis), which requires 10–12 hours and occupies about half of the cell-cycle time in a typical mammalian cell. After S phase, chromosome segregation and cell division occur in M phase (M for mitosis), which requires much less time (less than an hour in a mammalian cell). M phase comprises two major events: nuclear division, or mitosis, during which the copied chromosomes are distributed into a pair of daughter nuclei; and cytoplasmic division, or cytokinesis, when the cell itself divides in two
At the end of S phase, the DNA molecules in each pair of duplicated chromosomes are intertwined and held tightly together by specialized protein linkages. Early in mitosis at a stage called prophase, the two DNA molecules are gradually disentangled and condensed into pairs of rigid, compact rods called sister chromatids, which remain linked by sister-chromatid cohesion. When the nuclear envelope disassembles later in mitosis, the sister-chromatid pairs become attached to the mitotic spindle, a giant bipolar array of microtubules . Sister chromatids are attached to opposite poles of the spindle and, eventually, align at the spindle equator in a stage called metaphase. The destruction of sister-chromatid cohesion at the start of anaphase separates the sister chromatids, which are pulled to opposite poles of the spindle. The spindle is then disassembled, and the segregated chromosomes are packaged into separate nuclei at telophase. Cytokinesis then cleaves the cell in two, so that each daughter cell inherits one of the two nuclei.



The Eukaryotic Cell Cycle Usually Consists of Four Phases
Most cells require much more time to grow and double their mass of proteins and organelles than they require to duplicate their chromosomes and divide. Partly to allow time for growth, most cell cycles have gap phases—a G1 phase between M phase and S phase and a G2 phase between S phase and mitosis. Thus, the eukaryotic cell cycle is traditionally divided into four sequential phases: G1, S, G2, and M. G1, S, and G2 together are called interphase . In a typical human cell proliferating in culture, interphase might occupy 23 hours of a 24-hour cycle, with 1 hour for M phase. Cell growth occurs throughout the cell cycle, except during mitosis. The two gap phases are more than simple time delays to allow cell growth. They also provide time for the cell to monitor the internal and external environment. to ensure that conditions are suitable and preparations are complete before the cell commits itself to the major upheavals of S phase and mitosis. The G1 phase is especially important in this respect. Its length can vary greatly depending on external conditions and extracellular signals from other cells. If extracellular conditions are unfavorable, for example, cells delay progress through G1 and may even enter a specialized resting state known as G0 (G zero), in which they can remain for days, weeks, or even years before resuming proliferation. Indeed, many cells remain permanently in G0 until they or the organism dies. If extracellular conditions are favorable and signals to grow and divide are present, cells in early G1 or G0 progress through a commitment point near the end of G1 known as Start (in yeasts) or the restriction point (in mammalian cells). We will use the term Start for both yeast and animal cells. After passing this point, cells are committed to DNA replication, even if the extracellular signals that stimulate cell growth and division are removed.
Cell-Cycle Control Is Similar in All Eukaryotes
Some features of the cell cycle, including the time required to complete certain events, vary greatly from one cell type to another, even in the same organism. The basic organization of the cycle, however, is essentially the same in all eukaryotic cells, and all eukaryotes appear to use similar machinery and control mechanisms to drive and regulate cell-cycle events. The proteins of the cell-cycle control system have been so well conserved that many of them function perfectly when transferred from a human cell to a yeast cell. We can therefore study the cell cycle and its regulation in a variety of organisms and use the findings from all of them to assemble a unified picture of how eukaryotic cells divide. Several model organisms are used in the analysis of the eukaryotic cell cycle. The budding yeast Saccharomyces cerevisiae and the fission yeast Schizosaccharomyces pombe are simple eukaryotes in which powerful molecular and genetic approaches can be used to identify and characterize the genes and proteins that govern the fundamental features of cell division. The early embryos of certain animals, particularly those of the frog Xenopus laevis, are excellent tools for biochemical dissection of cell-cycle control mechanisms, while the fruit fly Drosophila melanogaster is useful for the genetic analysis of mechanisms underlying the control and coordination of cell growth and division in multicellular organisms. Cultured human cells provide an excellent system for the molecular and microscopic exploration of the complex processes by which our own cells divide.
Summary
Cell division usually begins with duplication of the cell’s contents, followed by distribution of those contents into two daughter cells. Chromosome duplication occurs during S phase of the cell cycle, whereas most other cell components are duplicated continuously throughout the cycle. During M phase, the replicated chromosomes are segregated into individual nuclei (mitosis), and the cell then splits in two (cytokinesis). S phase and M phase are usually separated by gap phases called G1 and G2, when various intracellular and extracellular signals regulate cell-cycle progression. Cell-cycle organization and control have been highly conserved during evolution, and studies in a wide range of systems have led to a unified view of eukaryotic cell-cycle control
1) http://tfscientist.hubpages.com/hub/Stages-of-the-Cell-Cycle-Mitosis-Part-2-of-2
2) http://foresight.org/Conference/MNT6/Abstracts/Knoch/index.html
Last edited by Admin on Tue Sep 11, 2018 8:53 pm; edited 14 times in total