https://reasonandscience.catsboard.com/t2017-the-amazing-organisation-and-design-of-dna-genomes-histones-nucleosomes-chromosomes
DNA, as a very stable nano-molecule, is an formidable, ideal massive storage device for long-term data archive. 22 The organisation of this higher order structure of chromatin and chromosomes is awe inspiring. We will have a closer look at it.
Eucaryotic DNA is Packaged into a set of Chromosomes
See the animatio below of the packing of DNA from double helix to chromosome
https://commons.wikimedia.org/wiki/File:DNA_packing.theora.ogv

The cells of the human body can produce at least 100,000 different types of proteins, all with a unique function. The information to make each of these complicated molecular machines is stored on the well-known molecule, DNA.
We think that we have done very well with human technology, packing information very densely on to computer hard drives, chips and CD-ROM disks. However, these all store information on the surface, whereas DNA stores it in three dimensions. It is by far the densest information storage mechanism known in the universe.
Let's look at the amount of information that could be contained in a pinhead volume of DNA. If all this information were written into paperback books, it would make a pile of such books 500 times higher than from here to the moon! The design of such an incredible system of information storage indicates a vastly intelligent Designer. 1
Eukaryotic chromosome structure 9
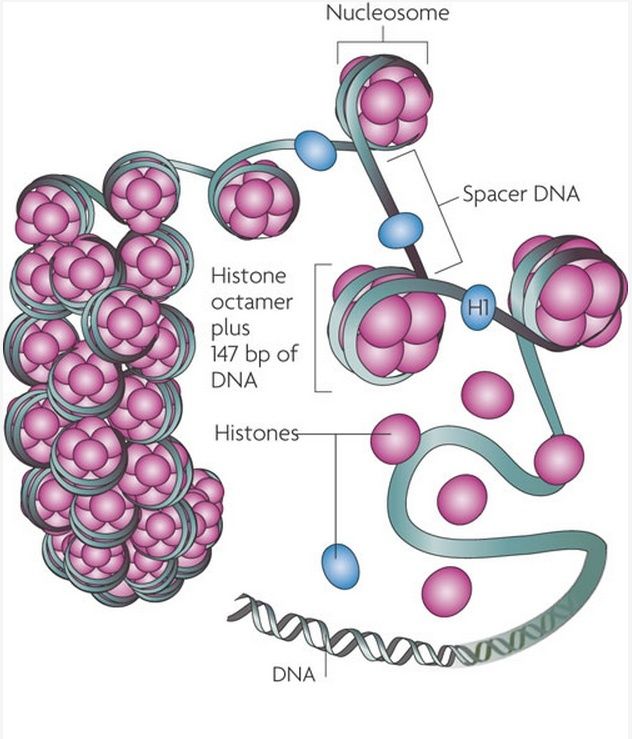
Packaging of DNA is facilitated by the electrostatic charge distribution: phosphate groups cause DNA to have a negative charge, whilst the histones are positively charged. Most eukaryotic cells contain histones (with a few exceptions) as well as the kingdom Archaea. Histones are positively charged molecules as they contain lysine and arginine in larger quantities and DNA is negatively charged. So they make a strong ionic bond in between them to form nucleosome.
Question : Are the right forces of ionic bonds through negative and positive charge not better explained through design, rather lucky trial and error processes ? Why should unguided processes produce such ingenious designs ?
Applied Intelligent Design: Storing Information on DNA
In terms of efficiency, DNA far surpasses any current manmade technology and can last for thousands of years. To get a handle on this, consider that 1 petabyte is equivalent to 1 million gigabytes of information storage. This paper reports an information storage density of 2.2 petabytes per gram.
The paper argues that since "existing technologies for copying DNA are highly efficient," this makes DNA an "excellent medium for the creation of copies of any archive for transportation, sharing or security." The authors conclude that "DNA-based storage has potential as a practical solution to the digital archiving problem and may become a cost-effective solution for rarely accessed archives." 19
Eukaryotic Chromosome Structure
Higher-order structure of chromatin and chromosomes
Packing ratio - the length of DNA divided by the length into which it is packaged
For example, the shortest human chromosome contains 4.6 x 107 bp of DNA (about 10 times the genome size of E. coli). This is equivalent to 14,000 µm of extended DNA, or about 2 mts. In its most condensed state during mitosis, the chromosome is about 2 µm long. This gives a packing ratio of 7000 (14,000/2).
To achieve the overall packing ratio, DNA is not packaged directly into final structure of chromatin. Instead, it contains several hierarchies of organization. The first level of packing is achieved by the winding of DNA around a protein core to produce a "bead-like" structure called a nucleosome. This gives a packing ratio of about 6. This structure is invariant in both the euchromatin and heterochromatin of all chromosomes. The second level of packing is the coiling of beads in a helical structure called the 30 nm fiber that is found in both interphase chromatin and mitotic chromosomes. This structure increases the packing ratio to about 40. The final packaging occurs when the fiber is organized in loops, scaffolds and domains that give a final packing ratio of about 1000 in interphase chromosomes and about 10,000 in mitotic chromosomes.
Eukaryotic chromosomes consist of a DNA-protein complex that is organized in a compact manner which permits the large amount of DNA to be stored in the nucleus of the cell. The subunit designation of the chromosome is chromatin. The fundamental unit of chromatin is the nucleosome. 20
Thats a amazing change , from a ratio of 6, to 10.000 !!
Squeezing DNA Into A Small Space 23
To fit 2 meters of DNA into a tiny nucleus is a monumental engineering feat. DNA is highly compacted yet has to be instantly available to rapidly make proteins in neurons with a momentary change of thought. This regulation is different in each type of cell. there are thousands of different types of neurons and each has different expression of the gene networks. It has been known for some time that the shape of proteins determines their function and the folding is very complex involving four levels of folding . Now it appears that the shape of the chromatin, also, determines function, with new secondary and tertiary structures discovered.
Condensins: universal organizers of chromosomes with diverse functions
Condensins are multisubunit protein complexes that play a fundamental role in the structural and functional organization of chromosomes in the three domains of life. 24It is a molecular machine that helps to condense and package chromosomes for cell replication. It is a five subunit complex, and is “the key molecular machine of chromosome condensation.

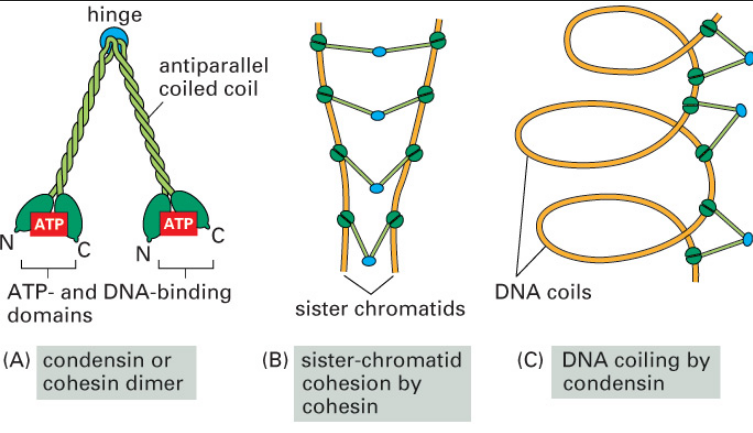

Chromatin
is a complex of macromolecules found in cells, consisting of DNA, protein and RNA. The primary functions of chromatin are
1) to package DNA into a smaller volume to fit in the cell,
2) to reinforce the DNA macromolecule to allow mitosis,
3) to prevent DNA damage, and 4) to control gene expression and DNA replication.
The primary protein components of chromatin are histones that compact the DNA. Chromatin is only found in eukaryotic cells (cells with defined nuclei). Prokaryotic cells have a different organization of their DNA (the prokaryotic chromosome equivalent is called genophore and is localized within the nucleoid region) 10
The structure of chromatin depends on several factors. The overall structure depends on the stage of the cell cycle. During interphase, the chromatin is structurally loose to allow access to RNA and DNA polymerases that transcribe and replicate the DNA. The local structure of chromatin during interphase depends on the genes present on the DNA: DNA coding genes that are actively transcribed ("turned on") are more loosely packaged and are found associated with RNA polymerases (referred to as euchromatin)
Everything in the cell is organized and in its expected place and function. Nothing in the cell is left to chance. The nucleus is no exception. In fact, in some ways, the nucleus is more organized and complex than the rest of the cell. One aspect of the complexity and organization of the nucleus is the chromatin. 13
FUNCTIONS
Packing and compacting DNA strands. If human DNA was stretched out, it would measure nearly 80 inches long. The primary function of chromatin is to take that 80 inches of DNA and coil and compact it enough to fit into an area no more that 0.0000004 inches in diameter in the cell nucleus. Not only does the chromatin have to coil and compact the DNA to fit into such a small area, but it also has to keep it all accessible and usable.
Nuclear organization. Certain sections of the genome are generally found in specific regions with the nucleus. The chromatin helps organize and compartmentalize the genome into their respective regions of the nucleus.
Strengthen DNA strands. During the metaphase stage of cell division, the structure of chromatin changes into a form whose function is geared primarily for strength in preventing damage to the DNA as the daughter chromosomes separate.
Replication and transcription during cell division. During the interphase stage of cell division, chromatin takes on two different forms: euchromatin and heterochromatin.
Euchromatin. A lightly packed form of chromatin that is often classified as containing active DNA. It contains a high concentration of genes and often associated with the transcription. That part of the euchromatin that is not transcribed usually is converted into heterochromatin.
Heterochromatin. A tightly packed form of chromatin that is often classified as containing inactive DNA and is involved in maintaining the integrity of chromosomes along with gene regulation. Heterochromatin is further divided into two types: constitutive and facultative. Constitutive heterochromatin contains regions of DNA that are not well expressed. They often are found near the centromere and telomere areas of the chromosome. Facultative heterochromatin is less consistent than constitutive heterochromatin in that it may exist in one region in one cell and not in the next. Facultative heterochromatin has been associated with such processes as morphogenesis or differentiation and may result in the silencing on an entire chromosome.
Chromatin is a complex molecular structure with multiple functions that seems perfectly designed to pack, strengthen and control DNA.
Chromatin Shapes
Chromatin is the word used for the large structure formed by the many nucleosomes. When chromatin was originally viewed under a microscope, two different kinds were called heterochromatin or euchromatin. But, in fact there are many subtypes.
Heterochromatin is highly compacted and is largely not active. It is localized at the edges of the nucleus. Despite early descriptions, it is actually at least five different states with different markings. It includes telomeres and centromeres. Constitutive heterochromatin is repetitive forming structures such as centromeres and telomeres. Facultative heterochromatin consists of genes that are suppressed and silenced by markings and RNA interference. It is not repetitive and can become active at some time.
Euchromatin is the active region and has a high density of genes, with RNAs and proteins. It is usually in the act of making proteins and is closer to the center of the nucleus.
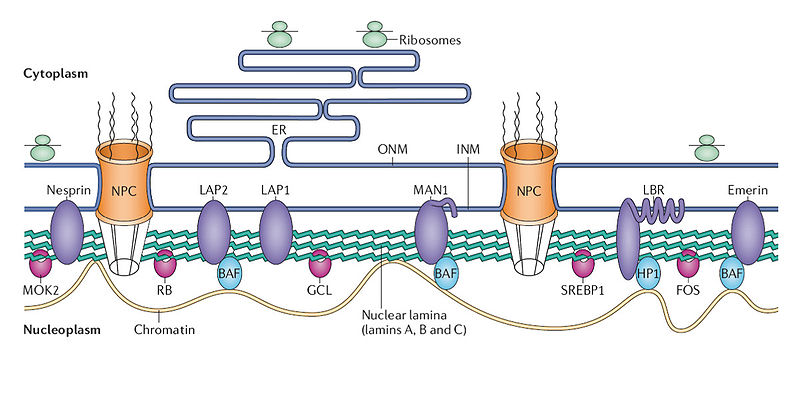
Nucleus Structure and Chromatin 23
The nucleus has a complex structure that is just now being discovered and the different types of chromatin fit in different compartments. The large nucleolus near the center has a membrane and its primary function is to synthesize and assemble ribosomes.
Complex structures near the edge of the nucleus are the nuclear lamina. The lamina are made of intermediate filaments (proteins called lamins) and proteins that are near or attached to the membrane. Lamins are a large family of proteins that form many very complex structures that are just being discovered. The lamina form compartments that organize the chromatin and influence replication and cell division. They bind specific chromatin through rod like structures to specific regions called matrix attachment regions. Lamina, also, bind to specific histones. The nuclear pores are complex structures in the nuclear membrane that determine what can come into the nucleus and what is sent out. The lamina are critical to the pore’s functions.
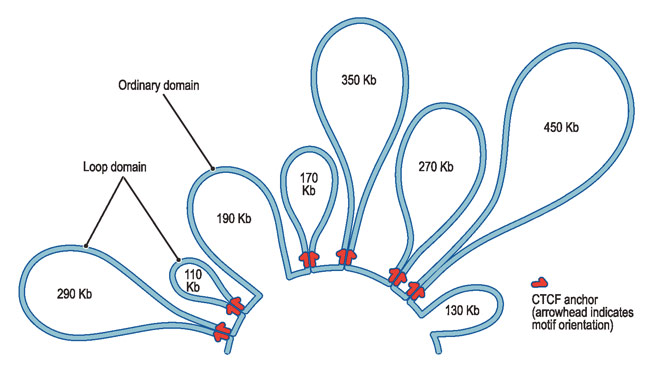
Loops of Chromatin
DNA loops HarvardThe first chromatin found to have an unusual looping 3D structure is a particular group of five genes making the beta subunit of hemoglobin. Genes sit on a loop right near the large regulator molecules needed to start and stop their production (promoters, enhancers and repressors). Loops can be flexible and the contact of the sites can be intermittent. This loop region makes it much easier to use the DNA. Often these loops create the environment for the activity, but a further stimulus is, also, needed.
Stem cells have been found to have less specific chromosome structures. As the cell differentiates into a specific cell type, then 3D structures appear limiting the cell’s function to the DNA regions that are forming loops. The new structure in the differentiated cell limits which genes are available—in essence defining the type of cell. Having pre formed structures makes the necessary proteins rapidly available by using these setups for close interaction of enhancers and promoters.
The bunching of nucleosomes has been termed “clutches,” for their comparison to the number of eggs that are left in a bird’s nest (called a clutch). The nucleosomes of the stem cells are much less densely packed with smaller clutches of nucleosomes. The more the packaging of the clutch, the more differentiated is the cell. The more the cell has capacity as a stem cell, the fewer nucleosomes are in the clutch.
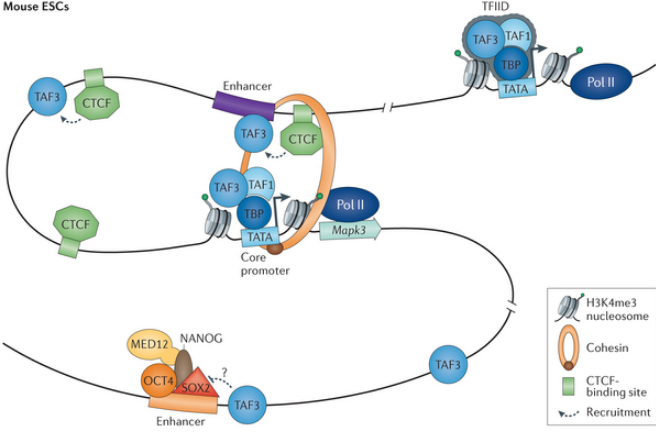
Protein Complexes Help Form 3D shape of Chromatin
Chromatin Remodeling in Eukaryotes 21
http://www.nature.com/scitable/topicpage/Chromatin-Remodeling-in-Eukaryotes-1082
chromatin not only serves as a way to condense DNA within the cellular nucleus, but also as a way to control how that DNA is used. In particular, within eukaryotes, specific genes are not expressed unless they can be accessed by RNA polymerase and proteins known as transcription factors. In its default state, the tight coiling that characterizes chromatin structure limits the access of these substances to eukaryotic DNA. Therefore, a cell's chromatin must "open" in order for gene expression to take place. This process of "opening" is called chromatin remodeling, and it is of vital importance to the proper functioning of all eukaryotic cells. In recent years, researchers have discovered a great deal about chromatin remodeling, including the roles that different protein complexes, histone variants, and biochemical modifications play in this process. However, a great deal remains to be learned before chromatin remodeling is fully understood.
Chromatin Remodeling at a Glance
Various molecules called chromatin remodelers provide the mechanism for modifying chromatin and allowing transcription signals to reach their destinations on the DNA strand. Understanding the nature and processes of these cellular construction workers remains an active area of discovery in genetic research.
Currently, investigators know that chromatin remodelers are large, multiprotein complexes that use the energy of ATP hydrolysis to mobilize and restructure nucleosomes. Recall that nucleosomes wrap 146 base pairs of DNA in approximately 1.7 turns around a histone-octamer disk, and the DNA inside each nucleosome is generally inaccessible to DNA-binding factors. Remodelers are thus necessary to provide access to the underlying DNA to enable transcription, chromatin assembly, DNA repair, and other processes. Just how remodelers convert the energy of ATP hydrolysis into mechanical force to mobilize the nucleosome, and how different remodeler complexes select which nucleosomes to move and restructure, remains unknown, however.
Remodelers are partitioned into five families, each with specialized biological roles. Nonetheless, all remodelers contain a subunit with a conserved ATPase domain. In addition to the conserved ATPase, each remodeler complex also possesses unique proteins that specialize it for its unique biological role. However, because all remodelers move nucleosomes and all such movement is ATP dependent, mobilization is most likely a property of the conserved ATPase subunit.
The ATPase domains of remodelers are similar in sequence and structure to known DNA-translocating proteins in viruses and bacteria. Recent evidence from the SWI/SNF and ISWI remodeler families has also revealed that remodeler ATPases are directional DNA translocases that are capable of the directional pumping of DNA. But how is this property applied to nucleosomes? It seems that the ATPase binds approximately 40 base pairs inside the nucleosome, from which location it pumps DNA around the histone-octamer surface. This enables the movement of the nucleosome along the DNA, thus permitting the exposure of the DNA to regulatory factors.
The additional domains and proteins that are attached to the ATPase are important for nucleosome selection, and they also help regulate ATPase activity. These attendant proteins bind to histones and nucleosomal DNA, and their binding to these molecules is affected by the histone modification state. The modification state helps determine whether the nucleosome is an appropriate substrate for a remodeler complex (Saha et al., 2006), as discussed later in this article.
Indeed, canonical histones can themselves be replaced by histone variants or modified by specific enzymes, thereby making the surrounding DNA more or less accessible to the transcriptional machinery.
So far, a number of histone variants have been found and localized to specific areas of chromatin. For instance, H2A.Z is a variant of H2A and is often enriched near relatively inactive gene promoters. Interestingly, H2A.Z does not take its place during replication when the chromatin structure is established. Instead, the chromatin remodeling complex SWR1 catalyzes an ATP-dependent exchange of H2A in the nucleosome for H2A.Z (Wu et al., 2005).
Histone Modification and the Histone Code
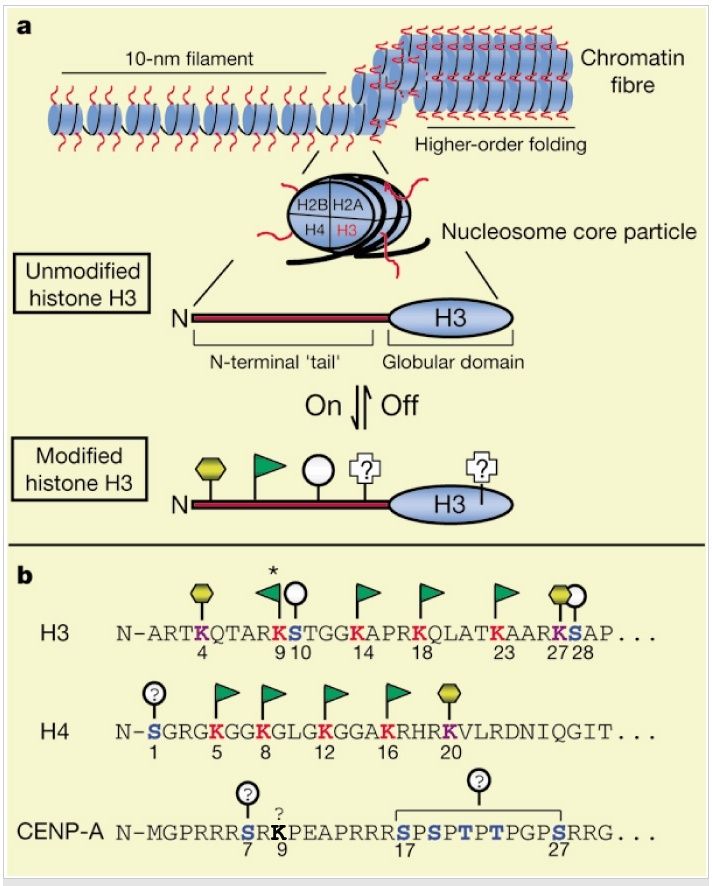
Histone sequences are highly conserved. The diagram below shows a typical chromatin fiber, with the blue cylinders representing histones. Extending from each of the histones is a "tail," called the N-terminal tail because proteins have two ends--an N terminus and C terminus. Here, the C terminus forms a globular domain that is packaged into the nucleosome. The other end of the histone is more flexible and capable of interacting more directly with DNA and the different proteins within the nucleus.
Figure below: (A) General chromatin organization. Like other histone "tails," the N terminus of H3 (red) represents a highly conserved domain that is likely to be exposed or extend outwards from the chromatin fiber. A number of distinct post-translational modifications are known to occur at the N terminus f H3 including acetylation (green flag), phosphorylation (grey circle), and methylation (yellow hexagon). Other modifications are known and may also occur in the globular domain. (B) The N terminus of human H3 is shown in single-letter amino-acid code. For comparison, the N termini of human CENP-A, a centromere-specific H3 variant, and human H4, the nucleosomal partner to H3, are shown. Note the regular spacing of acetylatable lysines (red), and potential phosphorylation (blue) and methylation (purple) sites. The asterisk indicates the lysine residue in H3 that is known to be targeted for acetylation as well as for methylation; lysine 9 in CENP-A (bold) may also be chemically modified (see text). The above depictions of chromatin structure and H3 are schematic; no attempt has been made to accurately portray these structures.
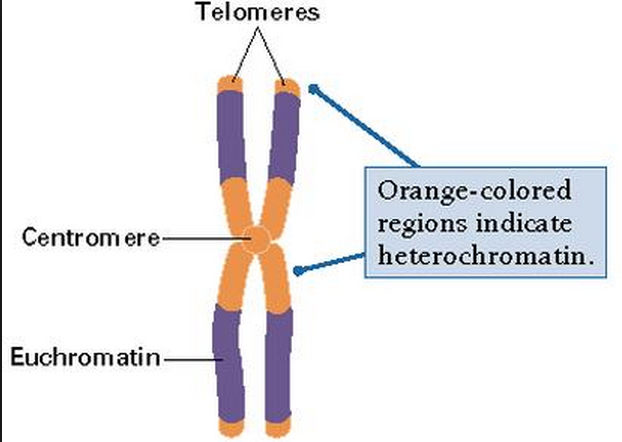
Euchromatin
is a lightly packed form of chromatin (DNA, RNA and protein) that is rich in gene concentration, and is often (but not always) under active transcription. Euchromatin comprises the most active portion of the genome within the cell nucleus. 92% of the human genome is euchromatic. The remainder is called heterochromatin. 12
Function
Euchromatin participates in the active transcription of DNA to mRNA products. The unfolded structure allows gene regulatory proteins and RNA polymerase complexes to bind to the DNA sequence, which can then initiate the transcription process. Not all euchromatin is necessarily transcribed, but in general that which is not is transformed into heterochromatin to protect the genes while they are not in use. There is therefore a direct link to how actively productive a cell is and the amount of euchromatin that can be found in its nucleus. It is thought that the cell uses transformation from euchromatin into heterochromatin as a method of controlling gene expression and replication, since such processes behave differently on densely compacted chromatin, known as the `accessibility hypothesis'. One example of constitutive euchromatin that is 'always turned on' is housekeeping genes, which code for the proteins needed for basic functions of cell survival.
 11
11
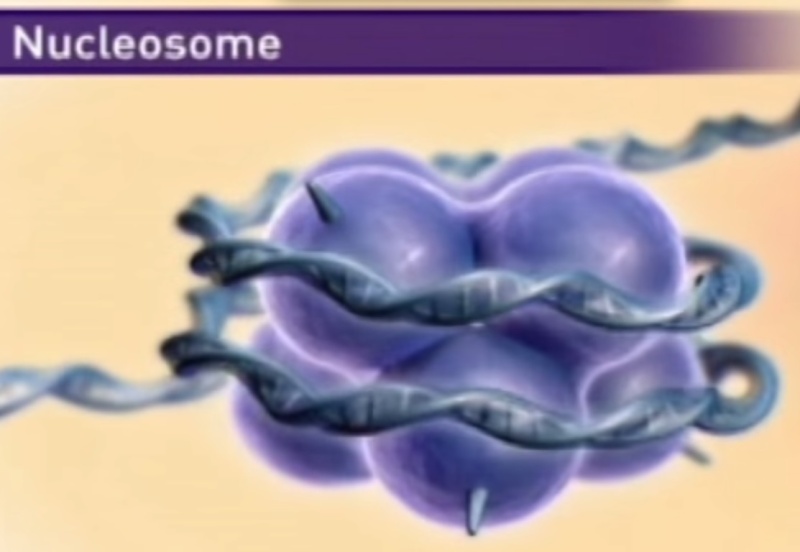
Nucleosome 14
A nucleosome is a basic unit of DNA packaging in eukaryotes, consisting of a segment of DNA wound in sequence around eight histone protein cores. This structure is often compared to thread wrapped around a spool.
Nucleosomes form the fundamental repeating units of eukaryotic chromatin, which is used to pack the large eukaryotic genomes into the nucleus while still ensuring appropriate access to it (in mammalian cells approximately 2 m of linear DNA have to be packed into a nucleus of roughly 10 µm diameter). Nucleosomes are folded through a series of successively higher order structures to eventually form a chromosome; this both compacts DNA and creates an added layer of regulatory control, which ensures correct gene expression. Nucleosomes are thought to carry epigenetically inherited information in the form of covalent modifications of their core histones.
“the information storage density of DNA, thanks largely to nucleosome spooling, is several trillion times that of the most advanced computer chips.” 15
So not only is there real information stored in DNA, but it is stored at a density on a molecular level, we can’t even approach with our best computers.
Dr. Stephen C. Meyer in his 1996 essay The Origin of Life and the Death of Materialism, wrote that
"the information storage density of DNA, thanks in part to nucleosome spooling, is several trillion times that of our most advanced computer chips 16
There are about 30 million nucleosomes in each human cell. So many are needed because the DNA strand wraps around each one only 1.65 times, in a twist containing 147 of its units, and the DNA molecule in a single chromosome can be up to 225 million units in length. 17
DNA has a striking property to pack itself in the appropriate solution conditions with the help of ions and other molecules. Usually, DNA condensation is defined as "the collapse of extended DNA chains into compact, orderly particles containing only one or a few molecules" 18
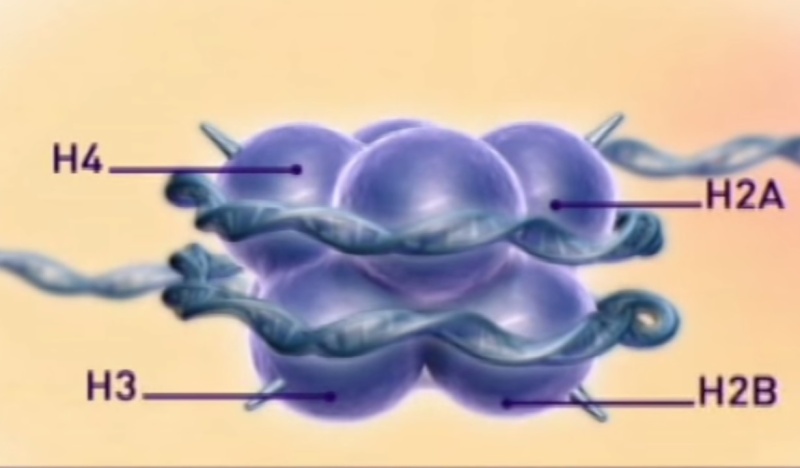
The basic level of DNA compaction is the nucleosome, where the double helix is wrapped around the histone octamer containing two copies of each histone H2A, H2B, H3 and H4. Linker histone H1 binds the DNA between nucleosomes and facilitates packaging of the 10 nm "beads on the string" nucleosomal chain into a more condensed 30 nm fiber. Most of the time, between cell divisions, chromatin is optimized to allow easy access of transcription factors to active genes, which are characterized by a less compact structure called euchromatin, and to alleviate protein access in more tightly packed regions called heterochromatin. During the cell division, chromatin compaction increases even more to form chromosomes, which can cope with large mechanical forces dragging them into each of the two daughter cells.

Histones:
In biology, histones are highly alkaline proteins found in eukaryotic cell nuclei that package and order the DNA into structural units called nucleosomes.They are the chief protein components of chromatin, acting as spools around which DNA winds, and play a role in gene regulation. Without histones, the unwound DNA in chromosomes would be very long (a length to width ratio of more than 10 million to 1 in human DNA). For example, each human cell has about 1.8 meters of DNA, (~6 ft) but wound on the histones it has about 90 micrometers (0.09 mm) of chromatin, which, when duplicated and condensed during mitosis, result in about 120 micrometers of chromosomes
Question: How did natural processes figure out to condense DNA into such a enormously tiny , highly regulated and functional structure ? Why at all should they do that ?
Histones are the major structural proteins of chromosomes. The DNA molecule is wrapped twice around a Histone Octamer to make a Nucleosome. Six Nucleosomes are assembled into a Solenoid in association with H1 histones. The solenoids are in turn coiled onto a Scaffold, which is futher coiled to make the chromosomal matrix.
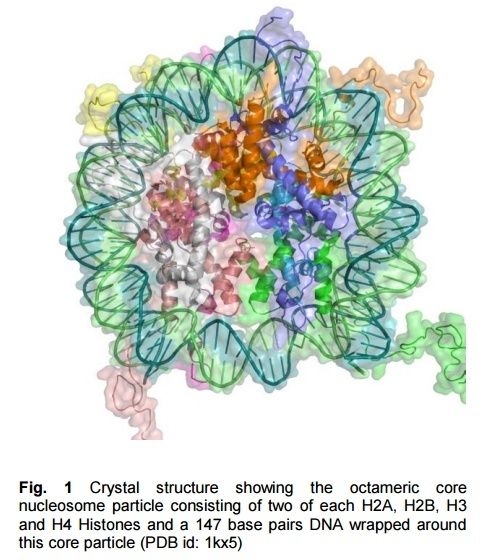
http://darwins-god.blogspot.com.br/2012/12/how-evolutionists-stole-histones.html
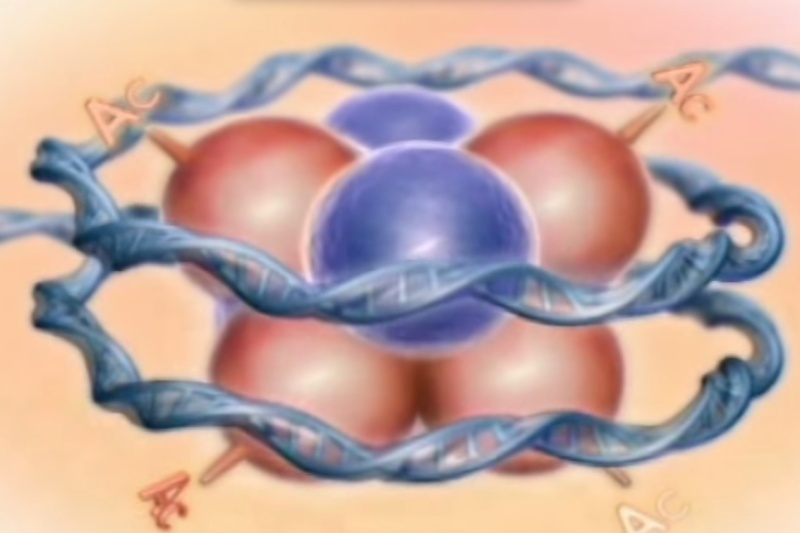

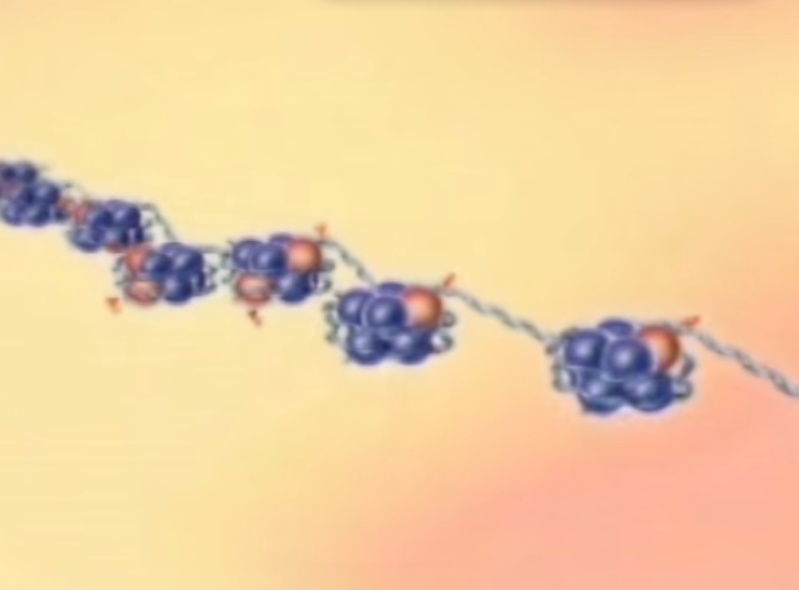
From molecular biology of the cell, B.Alberts, pg195 ( chapter 4 ):
As soon as genetics emerged as a science at the beginning of the twentieth
century, scientists became intrigued by the chemical structure of genes. The
information in genes is copied and transmitted from cell to daughter cell millions
of times during the life of a multicellular organism, and it survives the process
essentially unchanged.
What form of molecule could be capable of such
accurate and almost unlimited replication and also be able to direct the development
of an organism and the daily life of a cell? \A/hat kind of instructions does
the genetic information contain? How can the enormous amount of information
required for the development and maintenance of an organism fit within the
tiny space of a cell?
The answers to several of these questions began to emerge in the 1940s. At
this time, researchers discovered, from studies in simple fungi, that genetic
information consists primarily of instructions for making proteins. Proteins are
the macromolecules that perform most cell functions: they serve as building
blocks for cell structures and form the enzy'rnes that catalyze the cell's chemical
reactions
the hereditary information is carried on chromosomes,
threadlike structures in the nucleus of a eucaryotic cell
Chromosomes consist of both deoxyribonucleic acid (DNA) and proteins
the structure and chemical properties of DNA
make it ideally suited as the raw material of genes.
The packing has to be done in an orderly fashion so that the
chromosomes can be replicated and apportioned correctly between
the two daughter cells at each cell division
It must also allow access to chromosomal DNA for the enzymes that repair it when
it is damaged and for the specialized proteins that direct the expression of its
many genes.
CHROMOSOMAL DNA AND ITS PACKAGING IN THE CHROMATIN FIBER
The most important function of DNA is to carry genes, the information that
specifies all the proteins and RNA molecules that make up an organism including
information about when, in what types of cells, and in what quantity
each protein is to be made.
We also confront the serious challenge of DNA packaging. If the double
helices comprising all 46 chromosomes in a human cell could be laid end-toend,
they would reach approximately 2 meters; yet the nucleus, which contains
the DNA, is only about 6 pm in diameter.
This is geometrically equivalent to packing 40 km (24 miles) of extremely fine
thread into a tennis ball! The complex task of packaging DNA is accomplished by
specialized proteins that bind to and fold the DNA, generating a series of coils and
loops that provide increasingly higher levels of organization, preventing the DNA
from becoming an unmanageable tangle. Amazingly, although the DNA is very tightly
folded, it is compacted in a way that keeps it available to the many enzymes in
the cell that replicate it, repair it, and use its genes to produce RNA molecules
and proteins.
Following paper :
Integration of syntactic and semantic properties of the DNA code reveals chromosomes as thermodynamic machines converting energy into information 3
published online his summer is a true mind-blower showing the irreducible organizational complexity (author’s description) of DNA analog and digital information, that genes are not arbitrarily positioned on the chromosome etc.
The paper by Muskhelishvili and Travers, titled “Integration of syntactic and semantic properties of the DNA code reveals chromosomes as thermodynamic machines converting energy into information”,
argues that cellular mechanisms involved in processing genetic information make up an irreducibly complex system. The system requires genetic information, genetic machinery keyed to read that genetic information, as well as specific chromosomal organization. All of these components are necessary for what the paper calls "the organisational complexity of the genetic regulation system." 5
To be precise, the paper uses the term "irreducible organization" but it amounts to the same thing as biochemist Michael Behe's "irreducible complexity," and points implicitly to the same challenge to Darwinian accounts of origins.
the paper argues that in addition to the "digital information" in the primary DNA sequence, there is also "analog information" in the three-dimensional structure of chromosomes:
Recent studies have made it increasingly evident that the primary sequence of DNA in addition to the linear genetic code also provides three-dimensional information by means of spatially ordered supercoil structures relevant to all DNA transactions, including transcriptional control. In this review, we adopt the previously introduced terms "analog" and "digital" with regard to the two logically distinct types of information provided by the DNA. ... Any DNA gene is a carrier of digital information by virtue of its unique base sequence. Moreover, a gene conceived as an isolated piece of linear code (no matter whether this isolation occurs at the level of transcription or posttranscriptional processing), is a discontinuous entity that can be expressed or not, thus principally consistent with an "on-or-off" logic and, therefore, belonging to digital information type. Conversely, the physicochemical properties of DNA, as exemplified by supercoiling and mechanical stiffness, are determined not by individual base pairs but by the additive interactions of successive base steps. Supercoiling is by definition a continuous parameter ranging between positive and negative values (you can have more or less of it), and so belongs to analog information type.
The cell doesn’t need every protein encoded in its genome all the time. Sometimes genes are expressed (and the corresponding proteins are produced), and other times they aren’t. That is, the digital information in a gene can be expressed or not. To say it another way, the gene is a discontinuous entity that adheres to the “on-off” logic characteristic of digital information. 6
What hasn’t been apparent to biochemists is the analog information harbored in DNA—at least until Muskhelishvili and Travers reported it. They note that the nucleotide sequences of DNA not only encode information to make proteins, these sequences also impact the higher order architecture of DNA, which houses the analog information.
Most people recognize DNA’s iconic double helical structure, but what may not be well known is that the DNA double helix can adopt a variety of higher order shapes. One of the most prevalent architectures is referred to as a supercoil.
Supercoiling of DNA can be described by a number of parameters: (1) positive or negative; (2) twist; and (3) writhe. These parameters vary continuously, just like the grooves cut into a vinyl LP. In other words, supercoiling is an analog property.
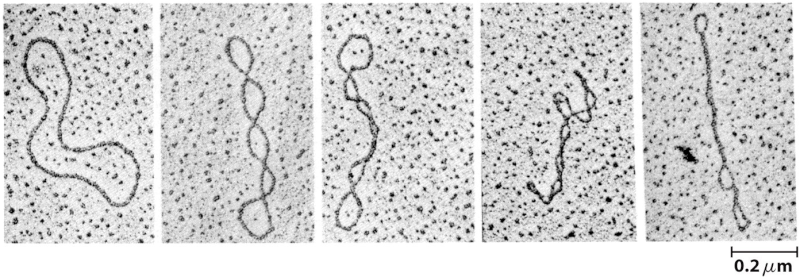
Below: Supercoiled (or "knotted"): Double stranded circular (or linear) DNA can have tertiary or higher order structure. Superhelicity is therefore sometimes referred to as DNA's tertiary structure. Supercoils refer to the DNA structure in which double-stranded circular DNA twists around each other. This is termed supercoiling, supertwisting or superhelicity -- meaning the coiling of a coil, also understood in terms of knots. Only topological closed domains (such as a covalently closed circle) can undergo supercoiling. A linear molecule can have topological domains as long as there is a region of the DNA bounded by constraints on the rotation of the DNA double helix. Eukaryotic DNAs in association with nuclear proteins acquire superhelical conformation in chromosomes 7
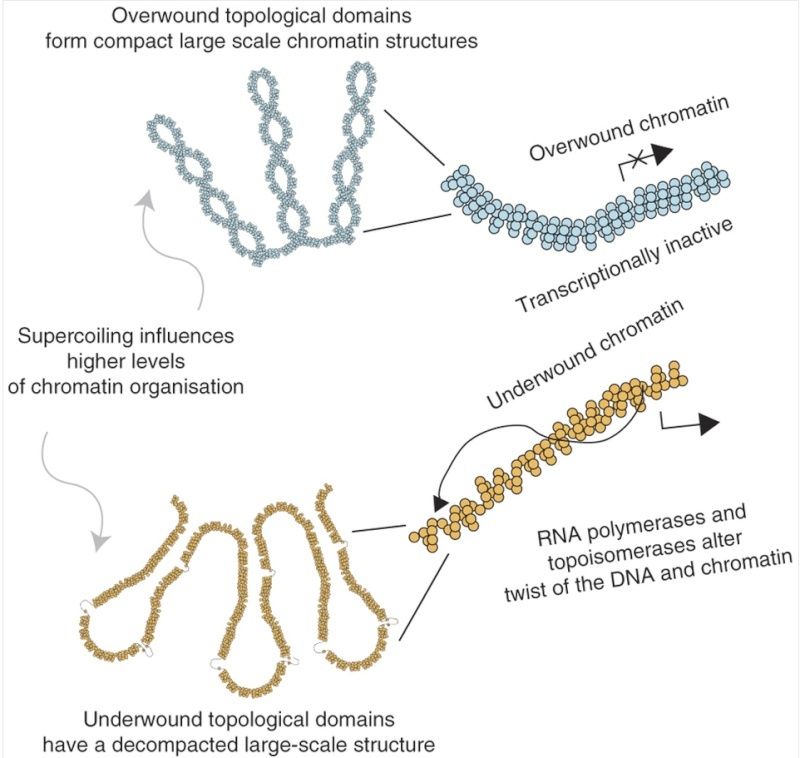
Below: Transcriptionally inactive chromatin is topologically overwound and has a cytologically compact large-scale chromatin structure. In contrast, a transcriptionally active region or transcriptional activation alters DNA supercoiling, remodeling supercoiling domains; this is accompanied by decompaction of large-scale chromatin structures. Therefore, large structural domains, for example as described by Hi-C31, are subdivided into smaller transcription-dependent supercoiling domains, providing an additional level of functional organization within the human genome. 8
As it turns out, the nucleotide sequence dictates the supercoiling parameters. Certain localized nucleotide sequences render some regions of the double helix more prone to supercoiling than others. On the other hand, some localized nucleotide sequences cause the DNA double helix to possess high flexibility, readily unwinding and untwisting.
Biochemists have come to realize that the higher order structures of DNA, and hence the analog information, play a significant role in gene expression, with the degree of supercoiling influencing the extent of gene expression.
The Interplay between DNA’s Digital and Analog Information
Muskhelishvili and Travers point out that the digital and analog information are coupled intrinsically through the nucleotide sequence. That is, the nucleotide sequence specifies the digital information of the gene and, at the same time, the higher order architectures of the genome, which in turn influence the expression of the digital information found in the gene. According to these two researchers, this coupling represents an “irreducible organizational complexity.”
They also note that proteins that interact with the genome aid in the coupling of digital and analog information. Proteins bind to specific nucleotide sequences. Once bound, these biomolecules help to promote supercoiling and stabilize higher order architectures. In other cases, bound proteins relax the supercoiling or destabilize the double helix. In other words, proteins play a role in regulating the expression of the digital information in the genome through the analog component.
The implications of this insight may be far reaching. Muskhelishvilli and Travers argue that it requires a new, more holistic strategy for studying gene expression, as opposed to the traditional approaches that treat gene regulation separately from the information found in the genes.
The paper then argues that the system of genetic regulation in cells is characterized by "irreducible organization" 5
Self-referential organisation, as we put it here, implies inter-conversion of information between logically distinct coding systems specifying each other reciprocally. Thus, the holistic approach assumes selfreferentiality (completeness of the contained information and full consistency of the different codes) as an irreducible organisational complexity of the genetic regulation system of any cell. Put another way, this implies that the structural dynamics of the chromosome must be fully convertible into its genetic expression and vice versa. Since the DNA is an essential carrier of genetic information, the fundamental question is how this self-referential organisation is encoded in the sequence of the DNA polymer.
The article even specifies that there are "Three basic components underlying the irreducible organisational complexity of any living cell" where "the organisation is essentially circular with all three basic components standing in relationship to reciprocal determination."
Those three components are specified as transcriptional machinery, DNA topology, and metabolic energy. The authors are perplexed by how the "irreducible" and "circular" organization of this system arose since they admit, "we face a 'chicken or egg' dilemma -- on the one hand the TF-TG interactions are determinative for the chromosomal structure, and on the other hand this very same structure determines the regulatory interactions."
What's incredible is that even though these two types of information are specified through different physical means, they nonetheless interact to regulate gene expression. The article explains that the supercoiling structure of DNA is vital to regulating gene expression, and at the same time it's not specified by the base-pair sequence. However, the base-pair sequence does interact with the supercoiling, and is more prone to localized untwisting to allow transcription.
How did these various independent levels of information become "coordinated"? Brilliance seems the best explanation for something brilliant.
Christianscientific adds following in regard of the paper :
It makes several very interesting points. First, the digital information of individual genes (semantics) is dependent on the the intergenic regions (as we know) which is like analog information (syntax). Both types of information are co-dependent and self-referential but you can’t get syntax from semantics. As the authors state, “thus the holistic approach assumes self-referentiality (completeness of the contained information and full consistency of the the different codes) as an irreducible organizational complexity of the genetic regulation system of any cell”. In short, the linear DNA sequence contains both types of information. Second, the paper links local DNA structure, to domains, to the overall chromosome configuration as a dynamic system keying off the metabolic signals of the cell. This implies that the position and organization of genes on the chromosome is not arbitrary. In other words, DNA topology (due to supercoiling and histone-like protein binding), Transcription, and Metabolic energy (ATP levels influence DNA gyrase activity, which affects supercoiling, which affects transcription) are all keying off each other and thus there is an overall order to the positioning of anabolic and catabolic genes relative to the origin of replication. In short, I think this is a fascinating review looking at DNA organization and function which, in the authors words, are irreducibly complex. 4
1) the authors are “serious” scientists, not fringe people
2) They are using “irreducible complexity” in the same sense as Behe. This is not a case of accidental use of the same phrase to mean something different. Their term “holistic” is another way of saying the same thing, that the system requires all of its parts to work.
3) This “holistic” approach is one that is becoming common in systems biology.
DNA - Replication, Wrapping & Mitosis
https://www.youtube.com/watch?v=Pj9cdVeIntY



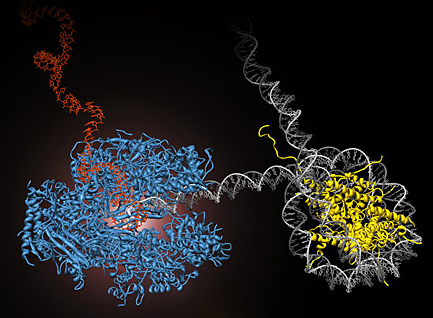
RNA polymerase II (blue) performs the first step of gene expression by moving along the cell’s DNA (gray) and transcribing it into messenger RNA (red). During this process, the polymerase encounters obstacles, such as nucleosomes, which tightly wrap the DNA around histone proteins (yellow) and prevent continued transcription. UC Berkeley researchers have developed methods to directly observe this process in real time. (Courtney Hodges/UC Berkeley)
1) http://reasonandscience.heavenforum.org/t1665-dna-information-storage?highlight=storage
2) http://www.berkeley.edu/news/media/releases/2009/07/30_nanomachines.shtml
3) http://link.springer.com/article/10.1007%2Fs00018-013-1394-1
4) http://www.christianscientific.org/refereed-scientific-article-on-dna-argues-for-irreducibly-complexity/
5) http://www.evolutionnews.org/2013/10/paper_irreducib077761.html
6) http://www.reasons.org/articles/digital-and-analog-information-housed-in-dna
7) http://www.udel.edu/chem/bahnson/chem645/websites/Sapra/Supercoiling.html
8 ) http://www.nature.com/nsmb/journal/v20/n3/fig_tab/nsmb.2509_F8.html
9) https://en.wikipedia.org/wiki/Eukaryotic_chromosome_structure
10) https://en.wikipedia.org/wiki/Chromatin
11) http://kc.njnu.edu.cn/swxbx/shuangyu/7.htm
12) https://en.wikipedia.org/wiki/Euchromatin
13) http://creationrevolution.com/chromatin-%E2%80%93-simple-cell-part-18/
14) http://en.wikipedia.org/wiki/Nucleosome
15) https://www.probe.org/mere-creation-science-faith-and-intelligent-design/
16) http://www.conservapedia.com/Creation_science
17) http://able2know.org/topic/72452-35
18) https://en.wikipedia.org/wiki/DNA_condensation
19) http://www.evolutionnews.org/2013/01/applied_intelli068631.html
20) http://www.ndsu.edu/pubweb/~mcclean/plsc431/eukarychrom/eukaryo3.htm
21) http://www.nature.com/scitable/topicpage/Chromatin-Remodeling-in-Eukaryotes-1082
22) http://www.ncbi.nlm.nih.gov/pmc/articles/PMC4222239/
23) http://jonlieffmd.com/blog/vast-complexity-of-chromatin-3d-shapes
24) http://genesdev.cshlp.org/content/26/15/1659.full
Last edited by Admin on Fri Jun 08, 2018 5:15 pm; edited 62 times in total


