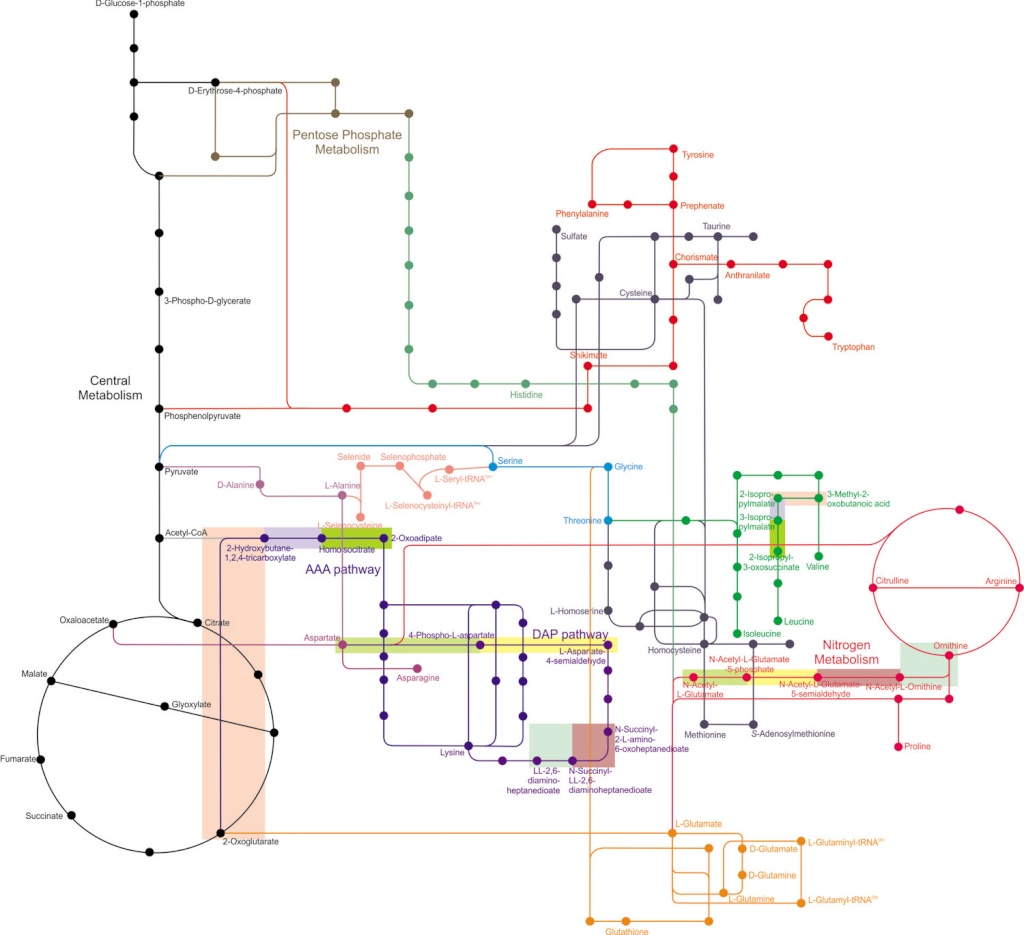
Major metabolic pathways and their inadequacy for origin of life proposals
https://reasonandscience.catsboard.com/t2004-major-metabolic-pathways-and-their-inadequacy-for-origin-of-life-proposals
According to geneticist Michael Denton, the break between the nonliving and the living world ‘represents the most dramatic and fundamental of all the discontinuities of nature.
And John Lennox writes in his book has science buried God ?
It is hard for us to get any kind of picture of the seething, dizzyingly complex activity that occurs inside a living cell, which contains within its lipid membrane maybe 100 million proteins of 20,000 different types and yet the whole cell is so tiny that a couple of hundred could be placed on the dot in this letter ‘i’.
The meaning of the genetic code is also virtually identical in all cells. The size, structure and component design of the protein synthetic machinery is practically the same in all cells. In terms of their basic biochemical design, therefore, no living system can be thought of as being primitive or ancestral with respect to any other system, nor is there the slightest empirical hint of an evolutionary sequence among all the incredibly diverse cells on earth.’
This view is supported by Nobel Prize-winner Jacques Monod, whom Denton cites. ‘We have no idea what the structure of a primitive cell might have been. The simplest living system known to us, the bacterial cell… in its overall chemical plan is the same as that of all other living beings. It employs the same genetic code and the same mechanism of translation as do, for example, human cells. Thus the simplest cells available to us for study have nothing “primitive” about them… no vestiges of truly primitive structures are discernible.’ Thus the cells themselves exhibit a similar kind of ‘stasis’ in connection with the fossil record.
Its interesting to try to figure out what that supposed last universal common ancestor ( LUCA ) was, in order to understand what kind of biochemical mechanisms, metabolism, enzymes, co-factors, proteins and genome information would have to be explained, and its origin.
From a biochemist’s perspective, life at the cellular level can be defined as a network of integrated and carefully regulated metabolic pathways, each contributing to the sum of activities that a cell must carry out. Cellular metabolism is a complex process involving about a thousand chemical reactions catalyzed by globular proteins, enzymes.
In the scientific paper: The Enzymatic and Metabolic Capabilities of Early Life, the author states that several independent studies have used comparative bioinformatics methods to identify taxonomically broad features of genomic sequence data, protein structure data, and metabolic pathway data in order to predict physiological features that were present in early, ancestral life forms. We survey modern metabolic pathways to identify those that maintain the highest frequency of metaconsensus enzymes. Using the full set of modern reactions catalyzed by these metaconsensus enzyme functions, we reconstruct a representative metabolic network that may reflect the core metabolism of early life forms.
Their research revealed the mind blowing complexity of Luca, and its metabolic pathways:
http://reasonandscience.heavenforum.org/t2174-the-enzymatic-and-metabolic-capabilities-of-early-life
According to another research paper : Evolution of the first metabolic cycles, There are two alternatives concerning the origin of life: the origin may be either heterotrophic or autotrophic. The paper : Analysis of the Intermediary Metabolism of a Reductive Chemoautotroph gives a idea of the complexity of it:
http://reasonandscience.heavenforum.org/t2147-the-naturalistic-approach-of-origin-of-life-scenarios
No wonder, do the authors of the paper: How Life Began: The Emergence of Sparse Metabolic Networks , openly admit that: ” The process by which the network of extant metabolism emerged is one of the major puzzles in the origin of life field.” Another paper admits that ” An open question for scientists is when and how cellular metabolism, the network of chemical reactions necessary to produce nucleic acids, amino acids and lipids, the building blocks of life, appeared on the scene.” The pathways for synthesis of most of the twenty amino acids used in proteins and the four nucleotides used in RNA are identical or nearly identical in Archaea, bacteria and eukaryotes, suggesting that these pathways were inherited from the LUCA. metabolic network. Thus, it appears that that the LUCA had the ability to synthesize the critical building blocks of life and did not rely on exogenous sources of these compounds. This supposition is supported by bioinformatic reconstructions of the genome of the LUCA. Biosynthetic pathways in extant organisms clearly resemble those in the LUCA. In the scientific paper : In The Ancient Ocean, Did Metabolism Precede The Origin Of Life?
http://reasonandscience.heavenforum.org/t2004-major-metabolic-pathways-and-their-inadequacy-for-origin-of-life-proposals
the author writes :
The observed chemical reactions occurred in the absence of enzymes but were made possible by the chemical molecules found in the Archean sea. Finding a series of reactions that resembles the “core of cellular metabolism” suggests that metabolism predates the origin of life. This implies that, at least initially, metabolism may not have been shaped by evolution but by molecules like RNA formed through the chemical conditions that prevailed in the earliest oceans.
Whether and how the first enzymes adopted the metal-catalyzed reactions described by the scientists remain to be established.
Its easily observable the hudge gap between the just so, almost helpless explanation attempts of the origin and arise of essential metabolic pathways, and their complexity observed even in the simplest cells.
This made the leading Origin of Life researcher Leslie Orgel say following:
The Implausibility of Metabolic Cycles on the Prebiotic Earth
Leslie E Orgel†
http://journals.plos.org/plosbiology/article?id=10.1371/journal.pbio.0060018
Almost all proposals of hypothetical metabolic cycles have recognized that each of the steps involved must occur rapidly enough for the cycle to be useful in the time available for its operation. It is always assumed that this condition is met, but in no case have persuasive supporting arguments been presented. Why should one believe that an ensemble of minerals that are capable of catalyzing each of the many steps of the reverse citric acid cycle was present anywhere on the primitive Earth, or that the cycle mysteriously organized itself topographically on a metal sulfide surface? The lack of a supporting background in chemistry is even more evident in proposals that metabolic cycles can evolve to “life-like” complexity. The most serious challenge to proponents of metabolic cycle theories—the problems presented by the lack of specificity of most nonenzymatic catalysts—has, in general, not been appreciated. If it has, it has been ignored. Theories of the origin of life based on metabolic cycles cannot be justified by the inadequacy of competing theories: they must stand on their own.
To accomplish any major task, a cell requires a series of reactions occurring in an ordered sequence. This, in turn, requires many different enzymes because most enzymes catalyze only a single reaction, and many such reactions are usually needed to accomplish a major biochemical operation. When we consider all the chemical reactions that occur within a cell, we are talking about metabolism (from the Greek word metaballein, meaning “to change”). The overall metabolism of a cell consists, in turn, of many specific metabolic pathways, each of which accomplishes a particular task. From a biochemist’s perspective, life at the cellular level can be defined as a network of integrated and carefully regulated metabolic pathways, each contributing to the sum of activities that a cell must carry out. Metabolic pathways are of two general types. Pathways that synthesize cellular components are called anabolic pathways (using the Greek prefix ana–, meaning “up”), whereas those involved in the breakdown of cellular constituents are called catabolic pathways (using the Greek prefix kata–, meaning “down”). Anabolic pathways usually involve a substantial increase in molecular order (and therefore a local decrease in entropy) and are endergonic (energy-requiring). Polymer synthesis and the biological reduction of carbon dioxide to sugar are examples of anabolic pathways. Often, anabolic pathways synthesize polymers such as starch and glycogen from glucose units in order to store energy for future use. Certain steroid hormones, for example, are called anabolic steroids because they stimulate the synthesis of muscle proteins from amino acids. Catabolic pathways, by contrast, are degradative pathways that typically involve a decrease in molecular order (increase in entropy) and are exergonic (energyliberating). These reactions often involve hydrolysis of macromolecules or biological oxidations. Catabolic pathways play two roles in cells: They release the free energy needed to drive cellular functions, and they give rise to the small organic molecules, or metabolites, that are the building blocks for biosynthesis. However, a catabolic pathway is not simply the reverse of the corresponding anabolic pathway. For example, the catabolic pathway for glucose degradation and the anabolic pathway for glucose synthesis use slightly different enzymes and intermediates. As we will see shortly, catabolism can be carried out either in the presence or absence of oxygen (i.e., under either aerobic or anaerobic conditions). The energy yield per glucose molecule is much greater in the presence of oxygen. However, anaerobic catabolism is also important, not only for organisms in environments that are always devoid of oxygen but also for organisms and cells that are temporarily deprived of oxygen.
http://www.jbc.org/content/278/48/47960.full
Cellular metabolism is a complex process involving about a thousand chemical reactions catalyzed by globular proteins, enzymes.

click image upload
A Theory of Biochemical Organization, Metabolic Pathways, and Evolution
http://citeseerx.ist.psu.edu/viewdoc/download?doi=10.1.1.464.3241&rep=rep1&type=pdf
http://www.jbc.org/content/278/48/47960.full
The Krebs Cycle
Whatever the metabolic specialization found in diverse living organisms (heterotrophy, photosynthetic autotrophy, chemosynthetic autotrophy, various forms of respiration and fermentation), there is a universal core of about 50 metabolic pathways involving the anabolism and catabolism of amino acids, fatty acids, saccharides (the glycolysis and the glycogenesis, the pentose phosphate pathway), and the Krebs cycle. Because the Krebs cycle is the point of confluence of all other metabolic pathways, it is sometimes viewed as primitive. That view is, however, challenged by several lines of evidence. Molecules entering the Krebs cycle (oxo acids and acyl-CoAs) are intermediate metabolites that are diversely interpreted with regard to their availability in primitive abiotic environments. Some biochemists consider them as most likely the products of a peripheral cellular metabolism. For example, the origin of the Krebs cycle was thought to be secondary and composite by Schoffeniels (1, 21), Gest (22, 23), and Meléndez-Hevia et al. (7). Others consider them as most likely available right from the beginning; for instance, carbon dioxide, water, and sunlight provide oxalic acid through pyrite catalysis. Then, the primitive soup would have been rich in dicarboxylic acids. These debates led us to incorporate two portions of the Krebs cycle as taxons in the matrix along with the metabolic pathways of each of the 16 aliphatic amino acids in our data sets. By analyzing anabolism and catabolism all together following the protocol of Cunchillos and Lecointre (24, 25) for catabolism only, we compared portions of the Krebs cycle both to amino acid catabolism and anabolism. The ultimate question was to find the earliest pathways and enzymatic functions among peripheral amino acid metabolisms and portions of the Krebs cycle.
http://www.evolutionnews.org/2008/02/leslie_orgel_metabolic_origin004792.html
The Implausibility of Metabolic Cycles on the Prebiotic Earth
Leslie E Orgel†
http://journals.plos.org/plosbiology/article?id=10.1371/journal.pbio.0060018
Almost all proposals of hypothetical metabolic cycles have recognized that each of the steps involved must occur rapidly enough for the cycle to be useful in the time available for its operation. It is always assumed that this condition is met, but in no case have persuasive supporting arguments been presented. Why should one believe that an ensemble of minerals that are capable of catalyzing each of the many steps of the reverse citric acid cycle was present anywhere on the primitive Earth, or that the cycle mysteriously organized itself topographically on a metal sulfide surface? The lack of a supporting background in chemistry is even more evident in proposals that metabolic cycles can evolve to “life-like” complexity. The most serious challenge to proponents of metabolic cycle theories—the problems presented by the lack of specificity of most nonenzymatic catalysts—has, in general, not been appreciated. If it has, it has been ignored. Theories of the origin of life based on metabolic cycles cannot be justified by the inadequacy of competing theories: they must stand on their own.
How Life Began: The Emergence of Sparse Metabolic Networks 1
The process by which the network of extant metabolism emerged is one of the major puzzles in the origin of life field. Metabolism may have been constructed only after the emergence of macromolecular RNAs (Joyce 2002), or a proto-metabolic network that generated amino acids and nucleotides (although possibly not ribonucleotides) may have set the stage for the emergence of the RNA World. These questions lie at the core of debates over the environment in which life arose, the source of the organic molecules that supplied the earliest form(s) of life, and the origin of extant metabolism. Thus, the core metabolome necessary to support life as we know it under the most demanding conditions (autotrophic growth) is an extremely sparse set of the possible small molecules.The pathways for synthesis of most of the twenty amino acids used in proteins and the four nucleotides used in RNA are identical or nearly identical in Archaea, bacteria and eukaryotes, suggesting that these pathways were inherited from the LUCA. The TCA cycle is present in representatives of all domains of life, although in some organisms only parts of the cycle are present, and in others the cycle runs in reverse. Note that there are cases in which non-homologous enzymes catalyze comparable reactions. For example, the glycolytic pathway in Archaea is similar to the glycolytic pathway in bacteria and eukaryotes, but a number of the Archaeal enzymes are not homologous to the analogous enzymes in bacteria and eukaryotes . Thus, it appears that that the LUCA had the ability to synthesize the critical building blocks of life and did not rely on exogenous sources of these compounds. This supposition is supported by bioinformatic reconstructions of the genome of the LUCA
The Critical Roles of Catalysts and Environmental Conditions in the Emergence of a Sparse Metabolic Network
Regardless of where life originated, the emergence of a sparse metabolic network depended critically on catalysis. The importance of rate acceleration by catalysts is obvious; nearly every reaction in extant cells is too slow to support life without a catalyst .
Does The Sparse Metabolic Network of Extant Life Resemble A Proto-Metabolic Network?
As discussed above, biosynthetic pathways in extant organisms clearly resemble those in the LUCA. A more difficult question is whether the structure of metabolism in the LUCA reflects the structure of a pre-existing proto-metabolic reaction network, or replaced a preexisting proto-metabolic reaction network. In the first scenario, metabolic pathways would have remained largely the same while more efficient catalysts were recruited to facilitate individual reactions, leading to a smooth transition from the earliest stages of mineral and small molecule catalysis, through an intermediate stage involving proto-RNA and RNA catalysts (likely with catalytic auxiliaries provided by amino acids, peptides, and cofactors), and finally to protein enzymes. A point in favor of this argument is that it is undoubtedly easier to patch a single catalyst into a functioning pathway than to invent de novo an entirely different pathway whose efficiency surpasses that of a previously existing pathway.
A second hypothesis is that modern metabolic pathways have completely replaced primordial pathways due to the advent of more effective catalysts, probably at the stage of the RNA World. Benner et al. (2009) have proposed that modern metabolism is a palimpsest and that re-writing of modern metabolism has largely obscured previously existing proto-metabolic pathways. This viewpoint is based upon the assumption that a large number of highly effective catalysts arose in the RNA World, or at least by the LUCA, which together allowed flux through pathways that had never before been accessible. In the context of Fig. 1, this would correspond to a switching between sets of catalysts with consequent reconstruction of the topology of the network.
The answer to this question most likely lies somewhere between these two opposing theories. The idea that modern metabolism runs along pathways that were laid down before the emergence of the LUCA is appealing from the standpoint of continuity between pre-life and life, and because recruitment of catalysts one at a time is more likely than recruitment of several catalysts simultaneously to enable an entirely new pathway. However, recruitment of several catalysts simultaneously to enable a novel pathway can certainly occur once there is a sufficient collection of catalysts
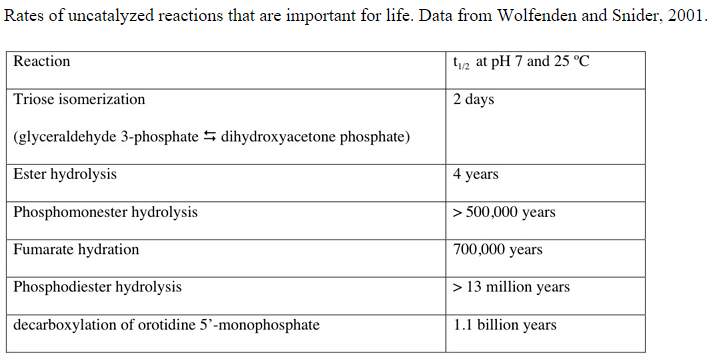
http://iose-gen.blogspot.com.br/2010/06/introduction-and-summary.html#methnat
The key to the functional complexity and specificity issue -- and identifying that "certain point" -- lies in Wicken's comparison to wiring diagrams for electrical circuits. For instance, we may compare (a) an outline map of a petroleum refinery with (b) the similar pathways of the living cell's metabolic reaction pathways:
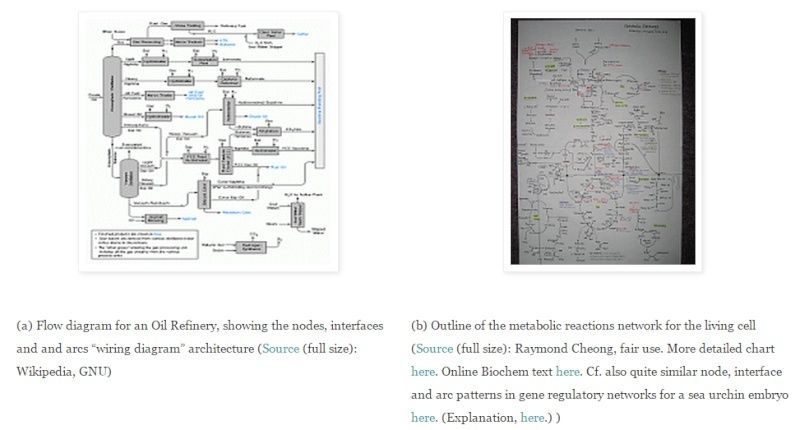
(This is a familiar exercise for anyone who has had to design, lay out, etch and then populate and solder together an electronics circuit board. [[A computer motherboard is a familiar example of such a circuit board.])
This “wiring diagram” approach is actually quite general purpose and powerful:
i --> The specific sequence of letters and numerical characters etc. in a word or sentence is such a network, usually called a string because the arcs and nodes form a linear sequence, like beads on a string: L-e-t-t-e-r-s.
ii --> In the refinery and metabolic networks just above, up to several arcs join to each node, and the whole forms an integrated network, perhaps something like a network of veins on a leaf or the mesh of lines and knots in a fishing net.
iii --> Perhaps surprisingly, we can model complex three-dimensional objects like the Old Man of the Mountain and the Mt Rushmore statues (or “exploded”- view diagrams of machines, etc.) as networks of connected points, sometimes called wire-frame models. Surface facets or skins and textures can be digitally draped over these “wire- frames.” The realism and specificity to a recognisable individual depend on how tightly spaced the network of fixed points is:
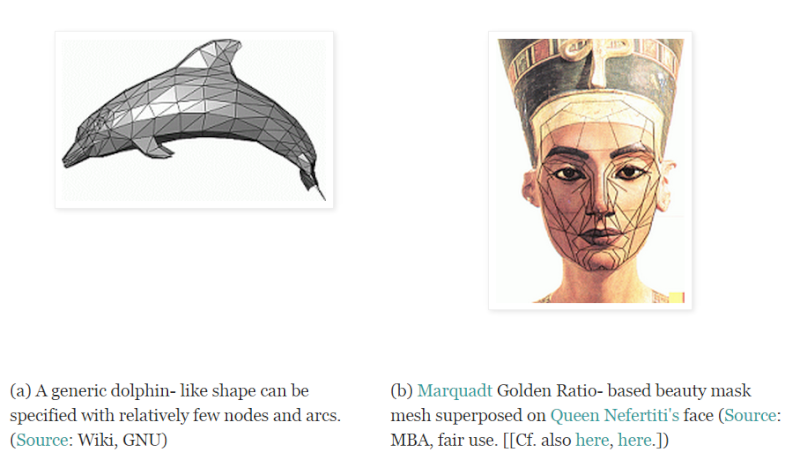
iv --> In each case, we can identify a chain of basic yes/no decisions or selections that specify the nodes [[which can be complex 3-D objects!], the connecting arcs and the ways the two interact. Then, we apply a rule of thumb: if the number of such basic yes/no decisions to build a sufficiently specific and functional network exceeds 500 - 1,000, we pass a reasonable threshold for “complexity.”
REASON: The number of possible configurations specified by 1,000 yes/no decisions, or 1,000 bits, is ~ 1.07 * 10^301; i.e. “roughly” 1 followed by 301 zeros. While, the ~ 10^80 atoms of the observed universe, changing state as fast as is reasonable [[the Planck time, i.e. every 5.39 *10^-44 s], for its estimated lifespan -- about fifty million times as long as the 13.7 billion years that are said to have elapsed since the big bang -- would only come up to about 10^150 states. Since 10^301 is ten times the square of this number, if the whole universe were to be viewed as a search engine, working for its entire lifetime, it could not scan through as much as 1 in 10^150 of the possible configurations for just 1,000 bits. That is, astonishingly, our “search” rounds down very nicely to zero: effectively no “search.” [[NB: 1,000 bits is routinely exceeded by the functionally specific information in relevant objects or features, but even so low a threshold is beyond the credible random search capacity of our cosmos, if it is not intelligently directed or constrained. That is, the pivotal issue is not incremental hill-climbing to optimal performance by natural selection among competing populations with already functional body forms. Such already begs the question of the need to first get to the shorelines of an island of specific function in the midst of an astronomically large sea of non-functional configurations; on forces of random chance plus blind mechanical necessity only. Cf. Abel on the Universal Plausibility Bound, here.]
Thirdly, causal factor (3) is about intelligence and its observable traces, not whether or not “supernatural” beings or immaterial minds actually exist and/or may act into our world. That is, the natural vs. supernatural contrast made by the NAS and Lewontin etc., is also a distractive strawman argument. Instead, a more appropriate contrast for scientific investigation on origins is: natural vs. ART-ificial (i.e. intelligent). Also, since we ourselves are intelligent, purposeful and inventive creatures – but cannot properly assume that we (or similar beings) are the only possible intelligences – we may easily see that intelligence is identifiable, recognisable and even definable as:
“. . . capacities to reason, to plan [[which entails (i) to purpose, (ii) to conceive or imagine a path to achieve it and (iii) to set out steps to fulfill the path], to solve problems, to think abstractly, to comprehend ideas, to use language, and to learn.” [[Wikipedia: article, “Intelligence.”]
Metabolic pathway maps
Carbon metabolism
Metabolic pathways Manet
http://en.wikipedia.org/wiki/Metabolic_pathway
1) http://cosmology.com/Abiogenesis113.html
https://reasonandscience.catsboard.com/t2004-major-metabolic-pathways-and-their-inadequacy-for-origin-of-life-proposals
According to geneticist Michael Denton, the break between the nonliving and the living world ‘represents the most dramatic and fundamental of all the discontinuities of nature.
And John Lennox writes in his book has science buried God ?
It is hard for us to get any kind of picture of the seething, dizzyingly complex activity that occurs inside a living cell, which contains within its lipid membrane maybe 100 million proteins of 20,000 different types and yet the whole cell is so tiny that a couple of hundred could be placed on the dot in this letter ‘i’.
The meaning of the genetic code is also virtually identical in all cells. The size, structure and component design of the protein synthetic machinery is practically the same in all cells. In terms of their basic biochemical design, therefore, no living system can be thought of as being primitive or ancestral with respect to any other system, nor is there the slightest empirical hint of an evolutionary sequence among all the incredibly diverse cells on earth.’
This view is supported by Nobel Prize-winner Jacques Monod, whom Denton cites. ‘We have no idea what the structure of a primitive cell might have been. The simplest living system known to us, the bacterial cell… in its overall chemical plan is the same as that of all other living beings. It employs the same genetic code and the same mechanism of translation as do, for example, human cells. Thus the simplest cells available to us for study have nothing “primitive” about them… no vestiges of truly primitive structures are discernible.’ Thus the cells themselves exhibit a similar kind of ‘stasis’ in connection with the fossil record.
Its interesting to try to figure out what that supposed last universal common ancestor ( LUCA ) was, in order to understand what kind of biochemical mechanisms, metabolism, enzymes, co-factors, proteins and genome information would have to be explained, and its origin.
From a biochemist’s perspective, life at the cellular level can be defined as a network of integrated and carefully regulated metabolic pathways, each contributing to the sum of activities that a cell must carry out. Cellular metabolism is a complex process involving about a thousand chemical reactions catalyzed by globular proteins, enzymes.
In the scientific paper: The Enzymatic and Metabolic Capabilities of Early Life, the author states that several independent studies have used comparative bioinformatics methods to identify taxonomically broad features of genomic sequence data, protein structure data, and metabolic pathway data in order to predict physiological features that were present in early, ancestral life forms. We survey modern metabolic pathways to identify those that maintain the highest frequency of metaconsensus enzymes. Using the full set of modern reactions catalyzed by these metaconsensus enzyme functions, we reconstruct a representative metabolic network that may reflect the core metabolism of early life forms.
Their research revealed the mind blowing complexity of Luca, and its metabolic pathways:
http://reasonandscience.heavenforum.org/t2174-the-enzymatic-and-metabolic-capabilities-of-early-life
According to another research paper : Evolution of the first metabolic cycles, There are two alternatives concerning the origin of life: the origin may be either heterotrophic or autotrophic. The paper : Analysis of the Intermediary Metabolism of a Reductive Chemoautotroph gives a idea of the complexity of it:
http://reasonandscience.heavenforum.org/t2147-the-naturalistic-approach-of-origin-of-life-scenarios
No wonder, do the authors of the paper: How Life Began: The Emergence of Sparse Metabolic Networks , openly admit that: ” The process by which the network of extant metabolism emerged is one of the major puzzles in the origin of life field.” Another paper admits that ” An open question for scientists is when and how cellular metabolism, the network of chemical reactions necessary to produce nucleic acids, amino acids and lipids, the building blocks of life, appeared on the scene.” The pathways for synthesis of most of the twenty amino acids used in proteins and the four nucleotides used in RNA are identical or nearly identical in Archaea, bacteria and eukaryotes, suggesting that these pathways were inherited from the LUCA. metabolic network. Thus, it appears that that the LUCA had the ability to synthesize the critical building blocks of life and did not rely on exogenous sources of these compounds. This supposition is supported by bioinformatic reconstructions of the genome of the LUCA. Biosynthetic pathways in extant organisms clearly resemble those in the LUCA. In the scientific paper : In The Ancient Ocean, Did Metabolism Precede The Origin Of Life?
http://reasonandscience.heavenforum.org/t2004-major-metabolic-pathways-and-their-inadequacy-for-origin-of-life-proposals
the author writes :
The observed chemical reactions occurred in the absence of enzymes but were made possible by the chemical molecules found in the Archean sea. Finding a series of reactions that resembles the “core of cellular metabolism” suggests that metabolism predates the origin of life. This implies that, at least initially, metabolism may not have been shaped by evolution but by molecules like RNA formed through the chemical conditions that prevailed in the earliest oceans.
Whether and how the first enzymes adopted the metal-catalyzed reactions described by the scientists remain to be established.
Its easily observable the hudge gap between the just so, almost helpless explanation attempts of the origin and arise of essential metabolic pathways, and their complexity observed even in the simplest cells.
This made the leading Origin of Life researcher Leslie Orgel say following:
The Implausibility of Metabolic Cycles on the Prebiotic Earth
Leslie E Orgel†
http://journals.plos.org/plosbiology/article?id=10.1371/journal.pbio.0060018
Almost all proposals of hypothetical metabolic cycles have recognized that each of the steps involved must occur rapidly enough for the cycle to be useful in the time available for its operation. It is always assumed that this condition is met, but in no case have persuasive supporting arguments been presented. Why should one believe that an ensemble of minerals that are capable of catalyzing each of the many steps of the reverse citric acid cycle was present anywhere on the primitive Earth, or that the cycle mysteriously organized itself topographically on a metal sulfide surface? The lack of a supporting background in chemistry is even more evident in proposals that metabolic cycles can evolve to “life-like” complexity. The most serious challenge to proponents of metabolic cycle theories—the problems presented by the lack of specificity of most nonenzymatic catalysts—has, in general, not been appreciated. If it has, it has been ignored. Theories of the origin of life based on metabolic cycles cannot be justified by the inadequacy of competing theories: they must stand on their own.
To accomplish any major task, a cell requires a series of reactions occurring in an ordered sequence. This, in turn, requires many different enzymes because most enzymes catalyze only a single reaction, and many such reactions are usually needed to accomplish a major biochemical operation. When we consider all the chemical reactions that occur within a cell, we are talking about metabolism (from the Greek word metaballein, meaning “to change”). The overall metabolism of a cell consists, in turn, of many specific metabolic pathways, each of which accomplishes a particular task. From a biochemist’s perspective, life at the cellular level can be defined as a network of integrated and carefully regulated metabolic pathways, each contributing to the sum of activities that a cell must carry out. Metabolic pathways are of two general types. Pathways that synthesize cellular components are called anabolic pathways (using the Greek prefix ana–, meaning “up”), whereas those involved in the breakdown of cellular constituents are called catabolic pathways (using the Greek prefix kata–, meaning “down”). Anabolic pathways usually involve a substantial increase in molecular order (and therefore a local decrease in entropy) and are endergonic (energy-requiring). Polymer synthesis and the biological reduction of carbon dioxide to sugar are examples of anabolic pathways. Often, anabolic pathways synthesize polymers such as starch and glycogen from glucose units in order to store energy for future use. Certain steroid hormones, for example, are called anabolic steroids because they stimulate the synthesis of muscle proteins from amino acids. Catabolic pathways, by contrast, are degradative pathways that typically involve a decrease in molecular order (increase in entropy) and are exergonic (energyliberating). These reactions often involve hydrolysis of macromolecules or biological oxidations. Catabolic pathways play two roles in cells: They release the free energy needed to drive cellular functions, and they give rise to the small organic molecules, or metabolites, that are the building blocks for biosynthesis. However, a catabolic pathway is not simply the reverse of the corresponding anabolic pathway. For example, the catabolic pathway for glucose degradation and the anabolic pathway for glucose synthesis use slightly different enzymes and intermediates. As we will see shortly, catabolism can be carried out either in the presence or absence of oxygen (i.e., under either aerobic or anaerobic conditions). The energy yield per glucose molecule is much greater in the presence of oxygen. However, anaerobic catabolism is also important, not only for organisms in environments that are always devoid of oxygen but also for organisms and cells that are temporarily deprived of oxygen.
http://www.jbc.org/content/278/48/47960.full
Cellular metabolism is a complex process involving about a thousand chemical reactions catalyzed by globular proteins, enzymes.

click image upload
A Theory of Biochemical Organization, Metabolic Pathways, and Evolution
http://citeseerx.ist.psu.edu/viewdoc/download?doi=10.1.1.464.3241&rep=rep1&type=pdf
http://www.jbc.org/content/278/48/47960.full
The Krebs Cycle
Whatever the metabolic specialization found in diverse living organisms (heterotrophy, photosynthetic autotrophy, chemosynthetic autotrophy, various forms of respiration and fermentation), there is a universal core of about 50 metabolic pathways involving the anabolism and catabolism of amino acids, fatty acids, saccharides (the glycolysis and the glycogenesis, the pentose phosphate pathway), and the Krebs cycle. Because the Krebs cycle is the point of confluence of all other metabolic pathways, it is sometimes viewed as primitive. That view is, however, challenged by several lines of evidence. Molecules entering the Krebs cycle (oxo acids and acyl-CoAs) are intermediate metabolites that are diversely interpreted with regard to their availability in primitive abiotic environments. Some biochemists consider them as most likely the products of a peripheral cellular metabolism. For example, the origin of the Krebs cycle was thought to be secondary and composite by Schoffeniels (1, 21), Gest (22, 23), and Meléndez-Hevia et al. (7). Others consider them as most likely available right from the beginning; for instance, carbon dioxide, water, and sunlight provide oxalic acid through pyrite catalysis. Then, the primitive soup would have been rich in dicarboxylic acids. These debates led us to incorporate two portions of the Krebs cycle as taxons in the matrix along with the metabolic pathways of each of the 16 aliphatic amino acids in our data sets. By analyzing anabolism and catabolism all together following the protocol of Cunchillos and Lecointre (24, 25) for catabolism only, we compared portions of the Krebs cycle both to amino acid catabolism and anabolism. The ultimate question was to find the earliest pathways and enzymatic functions among peripheral amino acid metabolisms and portions of the Krebs cycle.
http://www.evolutionnews.org/2008/02/leslie_orgel_metabolic_origin004792.html
The Implausibility of Metabolic Cycles on the Prebiotic Earth
Leslie E Orgel†
http://journals.plos.org/plosbiology/article?id=10.1371/journal.pbio.0060018
Almost all proposals of hypothetical metabolic cycles have recognized that each of the steps involved must occur rapidly enough for the cycle to be useful in the time available for its operation. It is always assumed that this condition is met, but in no case have persuasive supporting arguments been presented. Why should one believe that an ensemble of minerals that are capable of catalyzing each of the many steps of the reverse citric acid cycle was present anywhere on the primitive Earth, or that the cycle mysteriously organized itself topographically on a metal sulfide surface? The lack of a supporting background in chemistry is even more evident in proposals that metabolic cycles can evolve to “life-like” complexity. The most serious challenge to proponents of metabolic cycle theories—the problems presented by the lack of specificity of most nonenzymatic catalysts—has, in general, not been appreciated. If it has, it has been ignored. Theories of the origin of life based on metabolic cycles cannot be justified by the inadequacy of competing theories: they must stand on their own.
How Life Began: The Emergence of Sparse Metabolic Networks 1
The process by which the network of extant metabolism emerged is one of the major puzzles in the origin of life field. Metabolism may have been constructed only after the emergence of macromolecular RNAs (Joyce 2002), or a proto-metabolic network that generated amino acids and nucleotides (although possibly not ribonucleotides) may have set the stage for the emergence of the RNA World. These questions lie at the core of debates over the environment in which life arose, the source of the organic molecules that supplied the earliest form(s) of life, and the origin of extant metabolism. Thus, the core metabolome necessary to support life as we know it under the most demanding conditions (autotrophic growth) is an extremely sparse set of the possible small molecules.The pathways for synthesis of most of the twenty amino acids used in proteins and the four nucleotides used in RNA are identical or nearly identical in Archaea, bacteria and eukaryotes, suggesting that these pathways were inherited from the LUCA. The TCA cycle is present in representatives of all domains of life, although in some organisms only parts of the cycle are present, and in others the cycle runs in reverse. Note that there are cases in which non-homologous enzymes catalyze comparable reactions. For example, the glycolytic pathway in Archaea is similar to the glycolytic pathway in bacteria and eukaryotes, but a number of the Archaeal enzymes are not homologous to the analogous enzymes in bacteria and eukaryotes . Thus, it appears that that the LUCA had the ability to synthesize the critical building blocks of life and did not rely on exogenous sources of these compounds. This supposition is supported by bioinformatic reconstructions of the genome of the LUCA
The Critical Roles of Catalysts and Environmental Conditions in the Emergence of a Sparse Metabolic Network
Regardless of where life originated, the emergence of a sparse metabolic network depended critically on catalysis. The importance of rate acceleration by catalysts is obvious; nearly every reaction in extant cells is too slow to support life without a catalyst .
Does The Sparse Metabolic Network of Extant Life Resemble A Proto-Metabolic Network?
As discussed above, biosynthetic pathways in extant organisms clearly resemble those in the LUCA. A more difficult question is whether the structure of metabolism in the LUCA reflects the structure of a pre-existing proto-metabolic reaction network, or replaced a preexisting proto-metabolic reaction network. In the first scenario, metabolic pathways would have remained largely the same while more efficient catalysts were recruited to facilitate individual reactions, leading to a smooth transition from the earliest stages of mineral and small molecule catalysis, through an intermediate stage involving proto-RNA and RNA catalysts (likely with catalytic auxiliaries provided by amino acids, peptides, and cofactors), and finally to protein enzymes. A point in favor of this argument is that it is undoubtedly easier to patch a single catalyst into a functioning pathway than to invent de novo an entirely different pathway whose efficiency surpasses that of a previously existing pathway.
A second hypothesis is that modern metabolic pathways have completely replaced primordial pathways due to the advent of more effective catalysts, probably at the stage of the RNA World. Benner et al. (2009) have proposed that modern metabolism is a palimpsest and that re-writing of modern metabolism has largely obscured previously existing proto-metabolic pathways. This viewpoint is based upon the assumption that a large number of highly effective catalysts arose in the RNA World, or at least by the LUCA, which together allowed flux through pathways that had never before been accessible. In the context of Fig. 1, this would correspond to a switching between sets of catalysts with consequent reconstruction of the topology of the network.
The answer to this question most likely lies somewhere between these two opposing theories. The idea that modern metabolism runs along pathways that were laid down before the emergence of the LUCA is appealing from the standpoint of continuity between pre-life and life, and because recruitment of catalysts one at a time is more likely than recruitment of several catalysts simultaneously to enable an entirely new pathway. However, recruitment of several catalysts simultaneously to enable a novel pathway can certainly occur once there is a sufficient collection of catalysts

http://iose-gen.blogspot.com.br/2010/06/introduction-and-summary.html#methnat
The key to the functional complexity and specificity issue -- and identifying that "certain point" -- lies in Wicken's comparison to wiring diagrams for electrical circuits. For instance, we may compare (a) an outline map of a petroleum refinery with (b) the similar pathways of the living cell's metabolic reaction pathways:

(This is a familiar exercise for anyone who has had to design, lay out, etch and then populate and solder together an electronics circuit board. [[A computer motherboard is a familiar example of such a circuit board.])
This “wiring diagram” approach is actually quite general purpose and powerful:
i --> The specific sequence of letters and numerical characters etc. in a word or sentence is such a network, usually called a string because the arcs and nodes form a linear sequence, like beads on a string: L-e-t-t-e-r-s.
ii --> In the refinery and metabolic networks just above, up to several arcs join to each node, and the whole forms an integrated network, perhaps something like a network of veins on a leaf or the mesh of lines and knots in a fishing net.
iii --> Perhaps surprisingly, we can model complex three-dimensional objects like the Old Man of the Mountain and the Mt Rushmore statues (or “exploded”- view diagrams of machines, etc.) as networks of connected points, sometimes called wire-frame models. Surface facets or skins and textures can be digitally draped over these “wire- frames.” The realism and specificity to a recognisable individual depend on how tightly spaced the network of fixed points is:

iv --> In each case, we can identify a chain of basic yes/no decisions or selections that specify the nodes [[which can be complex 3-D objects!], the connecting arcs and the ways the two interact. Then, we apply a rule of thumb: if the number of such basic yes/no decisions to build a sufficiently specific and functional network exceeds 500 - 1,000, we pass a reasonable threshold for “complexity.”
REASON: The number of possible configurations specified by 1,000 yes/no decisions, or 1,000 bits, is ~ 1.07 * 10^301; i.e. “roughly” 1 followed by 301 zeros. While, the ~ 10^80 atoms of the observed universe, changing state as fast as is reasonable [[the Planck time, i.e. every 5.39 *10^-44 s], for its estimated lifespan -- about fifty million times as long as the 13.7 billion years that are said to have elapsed since the big bang -- would only come up to about 10^150 states. Since 10^301 is ten times the square of this number, if the whole universe were to be viewed as a search engine, working for its entire lifetime, it could not scan through as much as 1 in 10^150 of the possible configurations for just 1,000 bits. That is, astonishingly, our “search” rounds down very nicely to zero: effectively no “search.” [[NB: 1,000 bits is routinely exceeded by the functionally specific information in relevant objects or features, but even so low a threshold is beyond the credible random search capacity of our cosmos, if it is not intelligently directed or constrained. That is, the pivotal issue is not incremental hill-climbing to optimal performance by natural selection among competing populations with already functional body forms. Such already begs the question of the need to first get to the shorelines of an island of specific function in the midst of an astronomically large sea of non-functional configurations; on forces of random chance plus blind mechanical necessity only. Cf. Abel on the Universal Plausibility Bound, here.]
Thirdly, causal factor (3) is about intelligence and its observable traces, not whether or not “supernatural” beings or immaterial minds actually exist and/or may act into our world. That is, the natural vs. supernatural contrast made by the NAS and Lewontin etc., is also a distractive strawman argument. Instead, a more appropriate contrast for scientific investigation on origins is: natural vs. ART-ificial (i.e. intelligent). Also, since we ourselves are intelligent, purposeful and inventive creatures – but cannot properly assume that we (or similar beings) are the only possible intelligences – we may easily see that intelligence is identifiable, recognisable and even definable as:
“. . . capacities to reason, to plan [[which entails (i) to purpose, (ii) to conceive or imagine a path to achieve it and (iii) to set out steps to fulfill the path], to solve problems, to think abstractly, to comprehend ideas, to use language, and to learn.” [[Wikipedia: article, “Intelligence.”]
Metabolic pathway maps
Carbon metabolism
Metabolic pathways Manet
http://en.wikipedia.org/wiki/Metabolic_pathway
1) http://cosmology.com/Abiogenesis113.html
Last edited by Admin on Thu Feb 28, 2019 1:57 pm; edited 12 times in total