Major Supposed Differences between the LBCA and LUCA
1. Genetic Machinery of the Last Bacterial Common Ancestor (LBCA)
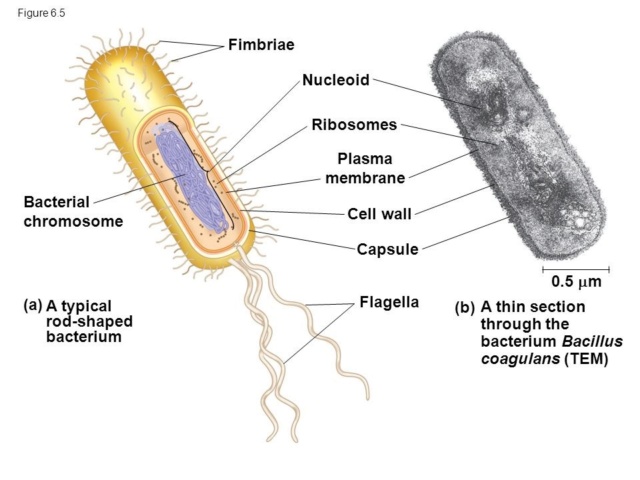
Transitioning our gaze from LUCA to the Last Bacterial Common Ancestor (LBCA), we begin to see evolutionary subtleties taking shape, representing an era of molecular innovation and divergence. LBCA, while descended from LUCA, would have possessed a slightly different toolkit, fine-tuned for its own unique existence in a constantly changing environment. At the heart of LBCA's machinery would have been the bacterial cell wall, a defining characteristic that separates it from its archaeal siblings. The machinery needed to synthesize peptidoglycan, the main component of this wall, would have emerged. Enzymes like transpeptidases and transglycosylases, acting as master architects, would carefully construct and maintain this protective barrier. The wall not only acts as a shield but also gives bacteria their distinct shapes - rods, spirals, and spheres. The advent of specialized RNA polymerases, distinct from those in LUCA, might have also made an appearance in LBCA, allowing for a more refined control over gene expression, in response to ever-changing environmental cues. Moreover, the evolution of various transport systems would have been essential. These molecular gatekeepers, ensuring the selective passage of nutrients and wastes, would be vital for LBCA's survival and efficiency. But what about the differences between LUCA and LBCA? One must consider the evolutionary trajectory, a series of steps and adaptations, leading from one to the other. This would involve the diversification and specialization of various proteins, adaptation to new ecological niches, and the establishment of unique metabolic pathways. The path from LUCA to LBCA would also have seen the evolution of sophisticated defense mechanisms against viruses and other threats, possibly including the earliest forms of bacterial immune systems, like the CRISPR-Cas system. To envision this journey from LUCA to LBCA is to embark on an odyssey of remarkable transformation. For LBCA to emerge from LUCA's shadows, it would have had to overcome countless challenges, each adaptation acting like a note in a grand evolutionary symphony. This narrative is not only a tale of biological innovation but also a testament to the tenacity and adaptability of life. It paints an enthralling story of evolution, filled with twists, turns, setbacks, and triumphs. And through this, it reminds us of the intricate beauty of life's molecular dance.
DNA replication
In the annals of Earth's ancient molecular history, LUCA, oft-pictured as a primordial beginner, instead strides forth as a maestro of DNA dynamics, its systems already a testament to eons of supposed prior refinement. Think of LUCA not as a novice pianist hitting the first tentative notes, but as a concert pianist seamlessly crafting symphonies. Its nucleic acid machinery, intricate and adept, would already be performing the dance of DNA replication and repair with finesse. LUCA's DNA polymerases, contrary to being rudimentary, would be akin to a composer's refined hands, effortlessly translating genetic scripts into harmonious strands of life. Helicases, those unsung heroes, would gracefully unzip the DNA strands, ensuring every note, every base pair, is accessible. Topoisomerases, the conductors of this genetic orchestra, would ensure the DNA's structure maintains its rhythm, neither too slack nor too taut. Yet, in the shadow of LUCA's prowess, the Last Bacterial Common Ancestor (LBCA) beckons, heralding a new age of nucleic sophistication. If LUCA was the master of a classical symphony, LBCA would be the vanguard of a new genre, blending tradition with innovation. LBCA's nucleic acid pathways would exemplify efficiency taken to new heights. Imagine LUCA's DNA replication as a grand river, flowing with purpose and precision. LBCA would introduce tributaries to this river, auxiliary pathways, which optimize nucleotide synthesis based on cellular cues and environmental dictates. The nucleotide synthesis, already a marvel in LUCA, would in LBCA, be a magnum opus of evolutionary engineering – faster, more efficient, and meticulously regulated. Maintenance and repair, too, would undergo a renaissance. In the theater of LBCA's cellular realm, the spotlight would shine on enhanced DNA repair mechanisms. Error correction would be near instantaneous, with an ensemble of enzymes orchestrating a ballet of precision repairs. The pathways, already established in LUCA, would now exhibit a kind of genetic agility in LBCA, swiftly adapting to diverse challenges, from UV radiation to chemical mutagens. Then, there's the art of DNA conservation, a realm where LBCA would truly shine. Picture a grand library in LUCA, storing vast tomes of genetic information. In LBCA, this library becomes alive, with "librarians" - enzymes, not just storing but also recycling, and rejuvenating old, damaged manuscripts into fresh scripts ready for a new audience. The ability to salvage and reuse nucleotides would be LBCA's testament to its commitment to sustainability and efficiency. In the continuum from LUCA to LBCA, we're invited to a concert that celebrates both tradition and innovation. From the established, masterful melodies of LUCA's nucleic realm, we transition to LBCA's reinventions, and its avant-garde approaches to synthesis, maintenance, and conservation. It's a tale of two maestros, each shaping the genetic orchestra of life in its unique, indelible way.
In the narrative of life's origin and progression, LUCA emerges not as an elemental prototype, but as a consummate maestro of molecular artistry, its very existence testifying to an ancient world steeped in complexity. This narrative raises profound questions about the plausibility of LUCA and its successors coming into being through simple, gradual processes. LUCA’s DNA polymerases, so vital for the dance of DNA synthesis and repair, can't be envisioned as primitive. They exhibit precision, seamlessly decoding genetic scripts into the harmonious strands of life. Yet, the question arises: How could such intricate machinery arise in small, beneficial increments? Even more perplexing is the interdependent dance of helicases and topoisomerases. Without one, the other's function is redundant, almost as if they were masterfully designed to work in harmony. As we trace the journey from LUCA to the Last Bacterial Common Ancestor (LBCA), the intricacies deepen. LBCA's genetic machinery evokes the image of a symphony, where each instrument is not just perfected, but also expertly fine-tuned to create an orchestrated masterpiece. Yet, how could such a system evolve through piecemeal, random mutations? The logic seems paradoxical. LBCA's heightened DNA repair mechanisms require a plethora of enzymes working together. The origin of each, in isolation, doesn't serve a functional purpose, making their independent, gradual evolution enigmatic. DNA conservation in LBCA takes the enigma to another level. The mechanism doesn’t just store information, but recycles and rejuvenates it. Without the entire system in place, partial mechanisms would be functionally obsolete. Such processes raise profound questions: How could intricate systems, where parts in isolation offer no advantage, arise through a step-by-step evolutionary path? The continuum from LUCA to LBCA illuminates a conundrum. The harmonious interplay of intricate systems, each dependent on the other, makes it challenging to envision their step-by-step, independent emergence. It's like expecting a symphony to arise from a sequence of random notes. The intricacies of molecular machinery, intertwined codes, precise signaling, and the sudden appearance of novel proteins all point towards an origin that isn't just the result of chance events but seems to resonate with purpose, precision, and design.
Molecular Components and Evolution: Transitioning from LUCA to LBCA implies the evolution of a myriad of molecular components. DNA polymerases would have to evolve from more basic structures in LUCA to more specialized versions in LBCA, facilitating more rapid and precise DNA synthesis. Helicases would undergo a transformation, perhaps increasing in efficiency or number, to ensure that DNA replication and transcription occur seamlessly.
Topoisomerases, given their role as 'conductors,' might have developed newer versions or auxiliary proteins to handle the increasingly complex DNA structure and dynamics. Novel regulatory elements would have to emerge, aiding in the nuanced control of gene expression as well as responding to external stimuli like UV radiation or chemical mutagens.
Integration of New and Evolving Parts: In the quantum leap from LUCA to LBCA, it's not just about the evolution of individual parts, but the harmonious integration of these parts into a more intricate system. The newly formed or evolved proteins would need to seamlessly integrate with the existing machinery. For example, if LBCA introduced novel helicases or topoisomerases, they'd need to work in tandem with the ancestral versions or replace them entirely without causing transcriptional chaos. New regulatory elements would need to establish connections with the existing genetic network, ensuring that any introduced control doesn't lead to cellular anarchy. This fine-tuning would involve complex feedback mechanisms to maintain homeostasis.
Challenges in Explaining the Transition: Accepting the transition from LUCA to LBCA solely based on unguided evolutionary mechanisms presents several challenges: The emergence of new proteins or regulatory elements poses a "chicken or the egg" conundrum. For instance, a novel regulatory element that controls a specific protein's expression is pointless without the protein's existence, and vice versa. The improbability lies in the spontaneous and concurrent emergence of complementary systems. One without the other renders them ineffective, making their simultaneous evolution a statistical challenge. The adaptation to diverse challenges like UV radiation implies the presence of a pre-existing mechanism to handle such adversities. An unguided evolution model would need to explain how such specific adaptations occurred in anticipation of future challenges. The enhanced DNA repair mechanisms of LBCA, as described, suggest almost instantaneous error correction. The evolution of such an efficient system, especially if LUCA's machinery was already adept, seems improbable without some form of guidance or extreme selective pressure. The evolution from LUCA to LBCA, as detailed in the narrative, implies not only the emergence of new molecular entities but also the harmonious orchestration of these components into a functional, efficient system. The challenges in explaining such a transition solely through unguided evolutionary mechanisms lie in the integration of these parts and the statistical improbabilities associated with the simultaneous evolution of complementary systems.
Transcription (from DNA to RNA)
If we now shift our focus from LUCA to the intricacies of transcription within the LBCA, we find a more refined symphony of cellular orchestration. Though the foundation of transcription was laid by LUCA, LBCA would have been a maestro of specialization, taking the baton and leading its molecular ensemble with precision. Within the LBCA's transcriptional machinery, the RNA Polymerase, while fundamentally similar, would have exhibited a greater degree of specialization. Tailored for bacterial needs, this enzyme would have been optimized for rapid response to environmental changes. Picture it: An RNA Polymerase, evolving in sophistication, more adeptly navigating the bacterial DNA, weaving RNA strands with heightened efficiency. Sigma factors would have played a pivotal role, introducing a layer of complexity and specificity to the transcription process in bacteria. Acting like meticulous conductors, these sigma factors would allow RNA Polymerase to recognize the precise starting point for transcription – the promoter regions. Each sigma factor would be specialized, responding to specific signals, ensuring that the genes essential for bacterial survival and thriving are transcribed precisely when needed. Additionally, the advent of bacterial operons, clusters of functionally related genes transcribed together as a single unit, would be a defining feature of the LBCA. This would represent a masterstroke in genetic efficiency, enabling coordinated gene expression in response to distinct environmental cues. The repressors and activators, acting as gatekeepers of these operons, would determine whether a gene cluster is activated or silenced, based on the cell's requirements. In this unfolding story of transcription from LUCA to LBCA, we would see an evolution from a foundational, yet rudimentary, system to a more intricate, specialized, and adaptive mechanism. It would be a testament to the marvel of ingenuity that evolution would require, where it would have the amazing ability of evolving the fine-tuning its processes, evolving not just to survive but to thrive, conducting an ever-complex molecular ballet in the theater of existence.
In the transition from LUCA to LBCA and the profound augmentation in transcriptional processes, one is left with the challenge of explaining the origination and precise integration of multiple components that seemingly push the boundaries of what unguided evolutionary mechanisms could achieve.
RNA Polymerase Specialization: While LUCA's RNA Polymerase set the stage for transcription, LBCA's version of this enzyme would need to be far more specialized. This means a plethora of specific modifications at the protein level for enhanced interaction with bacterial DNA, along with modifications for rapid response to environmental shifts. The question arises: How can random mutations lead to such nuanced, functional enhancements without causing detrimental effects in the process?
Emergence of Sigma Factors: The sigma factors are not mere accessories to RNA Polymerase; they are essential guides that ensure the enzyme knows where to start its transcription. Each sigma factor is tailored to recognize specific promoter regions. This demands the evolution of an entirely new set of proteins, each with a unique DNA-binding specificity. The precision required for this evolution, without causing mis-transcription or cellular chaos, poses a significant challenge to explain through mere chance.
Advent of Bacterial Operons: The clustering of functionally related genes to form operons is an exquisite feature of LBCA's transcription. Such a formation suggests a level of foresight: genes that need to be co-regulated end up adjacent on the genome. The probability of random mutations leading to this arrangement is hard to reconcile. Furthermore, the evolution of regulatory regions that can control these operons adds another layer of complexity.
Repressors and Activators: These aren't passive elements; they actively decide the fate of operons. This implies the emergence of a new set of proteins with specific binding capabilities, regulatory functions, and responsive mechanisms to environmental cues. The evolution of such proteins, in tandem with operons, suggests an intricacy hard to envision through random, stepwise mechanisms.
Integration and Interdependence: It's not just about the evolution of individual components but how they seamlessly integrate. RNA Polymerase's heightened function, the precise action of sigma factors, the strategic placement of genes in operons, and the nuanced control exerted by repressors and activators all converge into a finely-tuned system. The interdependence means that the malfunction or absence of one component could potentially disrupt the entire system.Given these considerations, the challenge lies not just in explaining the origin of individual parts, but in their integration into a coherent, highly efficient system. The precision, interdependence, and sophistication of the LBCA's transcriptional machinery defy the traditional model of gradual, unguided evolution. The intricate choreography of these molecular components seems to suggest a degree of orchestration that goes beyond mere evolution.
Translation (from RNA to Protein)
If we turn our gaze from the world of LUCA to the nuanced dance of translation in the LBCA, a more intricate and harmonious choreography comes into focus. While LUCA laid the foundational steps for translating the genetic code, the LBCA would have taken these moves and perfected them, introducing a series of sophisticated twirls and leaps to the routine. Within the heart of LBCA's translational arena, the ribosome might still hold center stage, but with marked differences. This ensemble of RNA and proteins, while reminiscent of its ancestral form in LUCA, would have been refined, with enhancements better suited to the bacterial way of life. Visualize a ribosome, with its evolutionary tweaks, more adeptly translating mRNA, and assembling proteins with unparalleled efficiency and precision. The dance partners of this ribosome, the tRNAs, would have displayed a level of specificity unparalleled in LUCA's world. Like accomplished dancers responding to nuanced cues, these tRNAs would have matched codons with elegance and accuracy, ensuring that each amino acid finds its rightful place in the emerging protein sequence. Aminoacyl-tRNA synthetases, ensuring the correct pairing of tRNAs and amino acids, would have acquired a discernment in LBCA, akin to master choreographers ensuring every move is perfectly executed. Their evolved precision would be critical, minimizing errors and allowing for the construction of more complex proteins vital to bacterial functions. The bacterial operon system, so central to transcription, would also have its echoes in translation. Imagine operons not just controlling when genes are transcribed, but also influencing how their resultant mRNAs are translated, coordinating the production of protein suites in response to environmental triggers. Yet, as in any intricate performance, there would be players ensuring that the dance goes smoothly. Molecular chaperones supposedly evolved in response to the LBCA's needs, would guide the nascent proteins, ensuring they fold into their correct shapes, ready to perform their functions. Moreover, regulatory proteins would have emerged, monitoring the translation process, ramping it up, or slowing it down in tune with the bacterial cell's needs. This transition from LUCA to LBCA, in the realm of translation, would depict an evolution from basic steps to an intricate ballet. Each adjustment, each new move, would symbolize the incredible potential that life would have needed to harness, orchestrating an elaborate dance of proteins in the theater of the microbial world.
Specific Molecular Components that Would Need to Emerge or Evolve
Ribosomal Enhancements: While both LUCA and LBCA would have utilized ribosomes, the LBCA's ribosomes would have showcased structural or functional enhancements to cater to the bacterial way of life. This might include specialized ribosomal proteins or rRNA variants.
Specialized tRNAs: With heightened specificity to ensure accurate translation, there would be a need for the evolution or diversification of tRNA molecules, each tailored to recognize distinct codons.
Aminoacyl-tRNA Synthetases: These enzymes attach the correct amino acid to its corresponding tRNA. Their evolution would mean a series of enzymes with refined specificity, reducing translation errors.
Molecular Chaperones: These would evolve to aid in the correct folding of proteins, ensuring that they adopt their functional conformations after translation.
Regulatory Proteins: New proteins or protein complexes that can modulate the speed and efficiency of the translation process would have emerged, allowing the cell to optimize protein synthesis according to its requirements.
Integration of these components to form a New Translational System
Coordinated Ribosome-tRNA Interplay: The enhanced ribosome, with its evolutionary tweaks, would need to seamlessly interact with the more specialized tRNAs, ensuring the ribosome's catalytic center (the peptidyl transferase center) functions optimally.
Aminoacyl-tRNA Synthetase-tRNA Partnership: Each synthetase would have to perfectly match with its corresponding tRNA, ensuring that amino acids are attached to the correct tRNAs without errors.
Operon System's Influence: In addition to their role in transcription, operons might have effects on translation, either directly or indirectly. This means coordination between transcriptional regulation and translational machinery.
Molecular Chaperone System: As proteins are synthesized, chaperones would need to interact with them without hindering the translation process, ensuring correct protein folding soon after synthesis.
Challenges or Improbabilities of this Transition through Unguided Evolutionary Mechanisms
Concurrent Evolution of Components: For translation to be efficient, several components (like tRNAs, synthetases, ribosomal units) would need to evolve concurrently. A refined tRNA is of little use without an equally refined aminoacyl-tRNA synthetase.
Integration Challenges: The emergence of new components doesn't guarantee their integration. For instance, a newly evolved regulatory protein would need a way to recognize and interact with the translational machinery effectively.
Error Minimization: The more complex the system, the greater the potential for errors. Evolutionary mechanisms would need to not only bring about new components but also ensure that the overall error rate in protein synthesis remains low.
Environmental Dependence: Given that LBCA would have faced different environmental challenges than LUCA, the evolution of its translational machinery would need to be finely tuned to these specific challenges, a feat difficult to achieve in a stepwise, unguided manner.
While the evolution from LUCA to LBCA showcases a remarkable expansion in translational complexity, there are profound challenges when considering how such intricate systems might have arisen solely through unguided processes. The complexity inherent in the transition from LUCA to LBCA is staggering, with a suite of intricate molecular components and subsystems needing to arise and integrate seamlessly. The realm of ribosomes provides a fitting starting point. For LBCA to function efficiently in its bacterial environment, its ribosomes would need to undergo significant enhancements. The emergence of new ribosomal proteins or rRNA variants is not a trivial matter. It’s not just about introducing new components but ensuring they work cohesively with the existing system. Further, consider the diversified tRNAs. It's not enough for them to simply exist; they must have exquisite specificity to ensure accurate translation. This raises an immediate question: In the absence of the aminoacyl-tRNA synthetases, what purpose would these refined tRNAs serve? The synthetases, in their role, would be redundant without the exact tRNAs they cater to. Now, factor in molecular chaperones. The complexity here is twofold. First, these molecules need to emerge. Second, they need to function in tandem with the translation process, ensuring proteins fold correctly immediately after synthesis. A mere emergence without synchronization would render them ineffective. The introduction of regulatory proteins, while crucial for the cell to modulate protein synthesis, presents another layer of complexity. These proteins must be not only structurally compatible with the existing system but must also carry out their function effectively. Their mere existence without functional utility would be purposeless.
Examining the challenges
Concurrent Evolution: The simultaneous evolution of interconnected components defies the traditional understanding of evolutionary progression. For instance, the need for a refined tRNA to evolve alongside its complementary aminoacyl-tRNA synthetase underscores a sophisticated level of coordination that seems improbable in a random, stepwise evolutionary process.
Integration Hurdles: The integration of new components is not guaranteed. Systems and molecules don't merely need to arise; they need to interface flawlessly with existing cellular machinery.
Error Minimization: The drive towards complexity necessitates an impeccable system of checks and balances. As the system grows intricate, the margin for error diminishes. A single misstep could be catastrophic, underscoring the necessity for a near-perfect system from inception.
Environmental Factors: The distinct environmental challenges faced by LBCA compared to LUCA means that the evolved components must not only be functionally adept but also environmentally robust. This fine-tuning presents yet another layer of improbability for a purely stepwise evolutionary progression.
Diving into the fabric of cellular function, we see codes, languages, and signaling mechanisms. This isn't just about individual systems; it's about the crosstalk, the communication, the intricate dance of molecular entities that must occur without a misstep. This seems to defy a piecemeal approach to its genesis. Instead, it resonates with the notion of a system designed with profound foresight, where each part is instantiated not in isolation but in perfect harmony with the whole. The suggestion that each of these components, each code, language, or signaling mechanism, emerged individually yet functioned collectively from the outset seems to transcend the bounds of traditional evolutionary paradigms.
Protein Folding and Post-translational Modifications
When we shift our focus from LUCA to the intricate artistry of protein folding and post-translational modifications within the LBCA, we're beckoned into a ballet of molecular grace and precision. In LUCA's realm, protein folding might have been a relatively straightforward process, driven mainly by intrinsic sequences and rudimentary environmental factors. One can picture proteins, freshly translated, cautiously finding their shape—guided predominantly by hydrophobic and hydrophilic interactions, reminiscent of a dancer tentatively learning the first steps. However, as we venture into the LBCA's world, the dance becomes infinitely more refined. Molecular chaperones, like seasoned dance instructors, would have emerged to assist in guiding the proteins. These chaperones, perhaps evolved versions of their precursors in LUCA, would play pivotal roles, ensuring that the proteins achieve their intended forms. Any misfolded proteins, akin to dancers missing a step, would either be corrected or marked for degradation, preserving the cellular harmony. Beyond just folding, the realm of post-translational modifications in the LBCA presents a spectacle of precision. If LUCA initiated the dance by introducing simple modifications, the LBCA would be where the true choreography emerges. Think of a suite of enzymes acting as backstage crew, each responsible for adding distinct embellishments—phosphates, methyl groups, or carbohydrates—to the protein performers. These modifications, much like costumes and makeup in a ballet, would not only enhance the appearance but also influence the performance, dictating the protein's function, location, or interactions within the bacterial cell. Furthermore, proteolytic cleavage, wherein proteins are strategically cut to activate or deactivate them, might have taken a center-stage role in the LBCA's repertoire. One can envision this like a dancer's pivotal spin or twist, altering the direction or tone of the entire performance. As we traverse from LUCA to LBCA, envision a transition from a basic dance school to a grand theater. The former teaches the essential steps, while the latter elevates them, adding layers of intricacy and meaning. The arena of protein folding and post-translational modifications would encapsulate this journey, highlighting the magnificent leaps and bounds life would have needed to make, orchestrating an exquisite molecular dance amidst the ever-evolving backdrop of existence.
In the time of LUCA, protein folding is thought to have been primarily influenced by basic environmental factors and the inherent sequences of amino acids. The proteins of this era were akin to tentative dancers, mostly relying on hydrophobic and hydrophilic interactions to attain their shapes. Transitioning towards the LBCA epoch, the introduction of molecular chaperones is posited. These entities, reminiscent of seasoned dance instructors, would guide proteins to their functional configurations, ensuring each performer fit seamlessly into the grand cellular ballet. LUCA's proteins, in their initial stages, would primarily have found their form with minimal assistance. The emergence of molecular chaperones by the LBCA's period suggests an enhanced level of sophistication in cellular processes. These chaperones, possibly evolving from rudimentary precursors, would be instrumental in preventing protein misfolding, ensuring that the cellular choreography remains harmonious. While the LUCA stage might have seen the beginning of simple protein modifications, it is within the LBCA's domain where the real spectacle is believed to have unfolded. A suite of enzymes, like a dedicated backstage crew, would introduce modifications such as phosphorylation, methylation, or glycosylation. These additions, analogous to costumes and makeup in a ballet, would modify protein functionality, localization, or cellular interactions. Within the LBCA’s framework, proteolytic cleavage becomes a central act, where proteins undergo strategic cuts either to activate or deactivate their functions. This process might be envisioned as a dancer's transformative spin, pivotal in altering the performance's direction or mood. As the intricacy of protein configurations elevated, the LBCA would necessitate refined quality control mechanisms. Misfolded proteins, like dancers missing their rhythm, would either be redirected to fold correctly or marked for degradation, ensuring the cellular ballet remains uninterrupted. Bridging the divide between LUCA and LBCA is the integrative evolution of these molecular processes. Each newly introduced or refined mechanism, from protein folding to quality control, would need to mesh seamlessly, ensuring that the grand molecular theater operates without a hitch.
Given the narrative of protein folding evolution from LUCA to LBCA, the leap from simple environmental influence to the existence of molecular chaperones appears immensely vast. For many proponents of Intelligent Design (ID), such complexity emerging within this evolutionary window suggests deliberate orchestration rather than the gradual progression of evolutionary forces. Molecular chaperones, acting as "dance instructors" for proteins, exhibit a level of specificity and purpose that seems to surpass mere chance. In the transition from LUCA's rudimentary protein folding mechanism to LBCA's sophisticated system with molecular chaperones, the absence of clear intermediate stages, or 'transitional forms', poses questions. The emergence of intricate molecular systems, such as the protein post-translational modifications and molecular chaperones, implies the addition of vast amounts of information within the cellular framework. The seamless integration of various cellular processes and systems, as evident in the transition from LUCA to LBCA, indicates a level of coherence that seems meticulously coordinated. Given the sheer complexity and specificity of molecular processes, the probability of them emerging spontaneously and functioning harmoniously becomes statistically challenging.
Nucleotide Synthesis and Recycling
Navigating from LUCA to the inner workings of the LBCA, we stumble upon a masterclass in efficiency and sustainability: the domain of nucleotide synthesis and recycling. In the earlier chapter of LUCA, nucleotide synthesis might have been a simpler narrative. The primordial pathways, relying on available substrates, would churn out the fundamental building blocks of life – adenine, guanine, cytosine, thymine, and uracil. Picture a nascent artisanal workshop, methodically crafting individual pieces, each nucleotide formed through basic but essential pathways, a true testament to nature's initial steps in crafting the genetic code. But as the curtain rises on LBCA's stage, we would have to witness a spectacle of evolved finesse and conservation. The pathways for nucleotide synthesis would have become streamlined, more adept, and efficient. Imagine a well-oiled assembly line, supposedly fine-tuned over eons, producing nucleotides with precision and speed tailored for the bacterial realm's exigencies. Yet, it's in the arena of nucleotide recycling where LBCA would truly have to demonstrate its virtuosity. The salvage pathways, akin to expert craftsmen repurposing materials, would ensure that no nucleotide goes to waste. Broken down DNA or RNA, rather than being discarded, would be a goldmine, their components recycled and refashioned into new nucleotides. This intricate dance of conservation and renewal, led by enzymes that would have perfected the art of salvage, would underscore LBCA's commitment to resource efficiency. Moreover, specialized bacterial enzymes would have had to emerge, optimizing the balance between de novo synthesis and recycling. These molecular maestros, sensing the cell's nucleotide pools, would either accelerate production or boost recycling, based on the cell's needs and environmental conditions. In this hypothesized evolutionary tale from LUCA to LBCA, we are invited to marvel at the huge steps in complexity and grace that would have to be achieved. The supposedly more rudimentary moves of nucleotide synthesis, despite already amazingly, unfathomably complex and orchestrated, initiated by LUCA, would have to be expanded upon, embellished, and refined by LBCA, introducing a symphony of even more capacity for recycling and conservation. This journey showcases a hypothetical glimpse into the mastery and intrinsic capacity that evolution would have to possess, where, faced with the challenges of existence, life would need to craft pathways that not only create but also conserve, dancing in harmonious rhythm with the ever-changing environment.
Starting with LUCA, the synthesis of nucleotides is thought to have depended heavily on available substrates. The basic building blocks – adenine, guanine, cytosine, thymine, and uracil – would emerge from these primordial processes. Come LBCA, these pathways would need to be far more efficient. The rudimentary systems of LUCA would have been replaced by streamlined pathways, built to rapidly and accurately produce nucleotides to sustain faster bacterial growth and replication. While LUCA might have functioned without an efficient system of recycling, LBCA would have needed to adopt salvage pathways. It's a system where degraded DNA or RNA wouldn't be considered waste. Instead, they would be invaluable resources. The components of these degraded nucleic acids would be refashioned into new nucleotides, ensuring maximum utilization of available resources. With the increasing complexity of nucleotide synthesis and recycling pathways, specialized enzymes would have been essential. These enzymes, tailored for the bacterial cell's requirements, would control and regulate the balance between de novo synthesis and nucleotide recycling. Acting like overseers, they would monitor nucleotide concentrations within the cell and modify metabolic pathways accordingly. With the vast intricacies in the nucleotide pathways, more complex regulatory mechanisms would have been necessitated. These mechanisms would act in tandem with the specialized enzymes, ensuring the optimal availability of nucleotides. They would consider factors like cellular demands, available substrates, and external environmental conditions. One of the most crucial aspects of the transition would be achieving a balance between producing new nucleotides and recycling old ones. While LUCA might have prioritized synthesis due to the abundance of available substrates, LBCA, in a possibly resource-limited environment, would need to ensure no wastage, emphasizing recycling without compromising on new synthesis. The evolutionary tale posits that both LUCA and LBCA, though separated by vast eons, would have faced their unique set of environmental and cellular challenges. While LUCA would have pioneered the foundational pathways, LBCA would have to refine them. It would be a dance of adaptation, where pathways would evolve in response to changing circumstances, ensuring the cell's survival and replication.
In the epoch of LUCA, the realm of nucleotide synthesis is imagined as a fledgling endeavor, grounded in its essential role but limited in its sophistication. Drawing from the substrates at its disposal, LUCA's metabolic mechanics would give rise to the foundational nucleotides: adenine, guanine, cytosine, thymine, and uracil. It was a straightforward process, seemingly archaic, but crucial for the genetic scripting of early life. Progressing towards the LBCA, the scenario changes dramatically. The envisioned pathways of nucleotide synthesis become subjects of rigorous refinement, evolving to resemble a meticulously coordinated production line, tailored to meet the voracious demands of bacterial propagation. A notable challenge in conceptualizing this evolutionary narrative is the transition from basic synthesis to the sophisticated recycling seen in the LBCA. The salvage pathways of the LBCA, with their ability to reclaim and repurpose components from degraded DNA or RNA, underscore the labyrinthine nature of this evolution. Such a transformative leap is not merely an enhancement of existing pathways; it implies the genesis of an entirely new system, one that meticulously reclaims cellular resources that LUCA would have potentially discarded. This complexity is further heightened when one considers the suite of enzymes that such a recycling mechanism would necessitate. The emergence of an array of novel enzymes, each tailored for specific roles in this recycling ballet, raises intriguing questions. These enzymes wouldn't function in isolation but would likely operate in tandem, where the product or by-product of one enzyme becomes the substrate for another. Such interdependent systems, while offering efficiency, also intensify the puzzle. How did these interlinked pathways, with enzymes reliant on one another, materialize in a coherent, functional order? Compounding this enigma is the postulated emergence of specialized bacterial enzymes in the LBCA. These aren't just mere catalysts; they're envisioned as regulators, arbiters of cellular balance. Tasked with the dual role of overseeing de novo nucleotide synthesis and the recycling processes, these enzymes would need to exhibit a heightened sensitivity to nucleotide concentrations, adjusting metabolic rates in real-time in response to the cell's internal and external milieu. In attempting to connect the dots from LUCA's rudimentary nucleotide synthesis to LBCA's intricate dance of synthesis and recycling, one ventures into a realm of profound metabolic evolution. The foundational pathways of LUCA, though monumental in their own right, would require not just augmentation but a radical overhaul to match the hypothesized sophistication of LBCA. Such an evolutionary narrative, teeming with the emergence of novel enzymes, intricate interdependencies, regulatory mechanisms, and feedback loops, poses substantial challenges. It compels one to ponder the depth and direction of the forces at play, and whether unguided mechanisms alone could sculpt such a masterpiece of metabolic complexity.
In light of the evidence and intricacies discussed pertaining to the hypothesized evolutionary transition from LUCA to LBCA, particularly in the realm of nucleotide synthesis and recycling, certain considerations lend weight to the polyphyletic creation perspective. Starting from the rudimentary operations in LUCA's time, the sheer intricacy and coordination required for the nuanced operations in LBCA's epoch is nothing short of staggering. The orchestrated dance of enzymes in LBCA, each with a distinct role yet interlinked in function, exhibits a level of interdependence and precision that is difficult to reconcile with incremental and unguided evolutionary steps. In such a scenario, a design perspective posits that these features and systems were intentionally encoded, reflecting an overarching intelligence rather than random mutations and natural selection alone. The sudden emergence of entirely new systems, particularly the salvage pathways and the regulatory enzymes specific to nucleotide metabolism, further complicate the evolutionary narrative. The genesis of these systems, especially in a coordinated and functional manner, demands explanations beyond mere beneficial mutations. The ID framework offers a vantage point where these systems are purposefully crafted and introduced, ensuring optimal efficiency and resource conservation. The interdependencies between enzymes, where the functionality of one is contingent on the presence or product of another, raise questions about their evolutionary origins. Such systems, where multiple parts must be simultaneously present and functional for the system to operate, challenge the gradualism inherent in classic evolutionary theory. A polyphyletic creation perspective interprets these interdependencies as evidence of intentional design, with each component crafted to fit within the larger system. The envisioned capabilities of LBCA, from its nucleotide synthesis to recycling, hint at a system that's not just functional but optimized. The balance between de novo synthesis and recycling, governed by specialized enzymes, showcases cellular machinery that's highly attuned to its environment and internal needs. Such optimality finds a more direct explanation in the ID paradigm, where systems are crafted for purpose and precision.
Repair and Protection mechanisms
Transiting from LUCA's primal world to the more evolved realm of the LBCA, one can't help but be fascinated by the escalating saga of DNA repair and protection mechanisms. In the rudimentary realm of LUCA, DNA repair might have had its nascent beginnings. Envision a lone artisan, painstakingly mending occasional tears, relying on basic tools and techniques. The mechanisms in place would have been essential, a primal response to ensure that the foundational story of life - embedded in DNA - remains untainted. Simple errors during replication or minor damages inflicted by environmental factors would be met with straightforward solutions. The mechanisms, while possibly efficient for the simpler life forms of that era, would be humble beginnings in the grand story of DNA repair and protection. However, as we lift the curtains on the world of LBCA, we would be faced with a veritable ballet of enhanced repair mechanisms, signifying a heightened recognition of DNA's importance. Picture a team of skilled craftsmen, each specializing in a different type of repair, equipped with tools sharpened and refined over time, supposedly through countless rounds of trial and error, mutations, and natural selection. Direct reversal of DNA damage, base excision repair, nucleotide excision repair, and mismatch repair - each pathway would have its own maestro, directing a coordinated response to specific types of damage. Moreover, in the face of threats that might cause double-strand breaks – possibly one of the most lethal forms of DNA damage – the LBCA would have to pioneer sophisticated repair strategies like homologous recombination or non-homologous end joining. These wouldn't just be simple fixes, but intricate processes ensuring genomic stability and integrity. Protection, on the other hand, would have been an equally captivating tale. If LUCA had a simple shield against environmental hazards, LBCA would have donned a full-fledged armor. This would include mechanisms to shield DNA from harmful radiation, chemicals, and reactive oxygen species. The emergence of DNA-binding proteins, acting like sentinels, would offer added layers of protection, ensuring the DNA remains coiled and guarded against potential threats. Navigating this supposed evolutionary leap from LUCA to LBCA, we'd see a transformation from basic repair and protection to an advanced, multi-layered defense system. This evolutionary narrative would require nature to repeatedly and intricately refine its tools, creating a system not just for survival, but to safeguard the specialized advanced molecular machinery of bacteria. Through this lens, we're invited to ponder the supposed capabilities and intricacies that evolution would need to navigate, ensuring life's dance remains uninterrupted in the theater of existence.
In the context of DNA repair and protection mechanisms, the evolutionary transition from LUCA to LBCA can be narrated as an amplification of intricacy and specificity.
1. Evolution of DNA Repair Mechanisms
a. Direct reversal of DNA damage
Initial Mechanism: Simple enzymatic reactions that might reverse minor chemical modifications in DNA.
LBCA Transition: Specialized enzymes, such as photolyases, would have evolved that can utilize light energy to directly reverse UV-induced DNA damage like pyrimidine dimers.
b. Base Excision Repair (BER)
Initial Mechanism: Rudimentary enzymes that recognize and cut out damaged or incorrect bases.
LBCA Transition: A suite of glycosylases would have emerged, each specific to a different kind of base lesion. Further, enzymes like AP endonucleases would have evolved to process the resulting abasic site.
c. Nucleotide Excision Repair (NER)
Initial Mechanism: Simple protein machinery recognizing bulky DNA adducts.
LBCA Transition: Complex protein complexes, like the UvrABC system, would be essential to recognize, incise, and replace a segment of DNA containing the lesion.
d. Mismatch Repair (MMR)
Initial Mechanism: Basic proofreading abilities of primitive DNA polymerases.
LBCA Transition: A sophisticated system comprising proteins like MutS, MutL, and MutH would have been essential to recognize and repair mismatched bases that escape the proofreading ability of DNA polymerase.
e. Double-strand Break Repair
Initial Mechanism: Possibly none, as double-strand breaks in LUCA's realm could have been lethal.
LBCA Transition: Two main pathways would emerge:
Homologous recombination, where a sister chromatid serves as a template for repair. Non-homologous end joining, directly ligating the broken DNA ends together.
2. DNA Protection Mechanisms
a. Protection against environmental hazards
Initial Mechanism: Simple DNA-binding proteins that might shield DNA from chemical or physical agents.
LBCA Transition: The appearance of specialized DNA-binding proteins that not only protect DNA but also regulate its topology, like histone-like proteins in bacteria.
b. Protection against Reactive Oxygen Species (ROS)
Initial Mechanism: Primitive antioxidants or enzymes detoxifying ROS.
LBCA Transition: Advanced antioxidant systems and enzymes like superoxide dismutase, catalase, and peroxidases would emerge to neutralize various ROS, protecting DNA from oxidative damage.
3. DNA Packing and Structuring
a. Initial DNA structuring
Initial Mechanism: Simple coiling of DNA or rudimentary binding proteins to compact DNA.
LBCA Transition: Introduction of specialized proteins and structures, such as the bacterial nucleoid-associated proteins, to compact, structure, and organize the bacterial chromosome efficiently.
In this speculative narrative, the transition from LUCA to LBCA embodies an evolution from simplicity to complexity. The DNA repair and protection mechanisms, believed to be rudimentary during LUCA's era, would undergo multiple stages of refinements to yield the advanced systems in the LBCA, underscoring the adaptability and resilience of life in the face of genomic challenges.
The transition from the rudimentary DNA repair and protection mechanisms of LUCA to the sophisticated systems found in the LBCA presents a series of challenges when considered through the lens of unguided evolutionary mechanisms. The evolution of direct reversal of DNA damage, for instance, would necessitate the emergence of enzymes like photolyases, which can harness light energy to reverse specific DNA lesions. The development of such specialized enzymes requires a series of coordinated mutations. The specificity and efficiency of these enzymes, especially in the context of a vast chemical landscape of potential DNA modifications, is remarkable. Moreover, base excision repair in the LBCA would demand a suite of glycosylases, each tailored to recognize a different kind of DNA lesion. The specificity of these enzymes is crucial; any error in recognition or processing could result in further genomic instability. The evolution of such precision, especially without a guiding mechanism, raises questions. For nucleotide excision repair, the emergence of complex protein complexes like the UvrABC system is crucial. These proteins must work in a coordinated fashion, each recognizing and processing a specific step in the repair pathway. The evolution of such a coordinated system, where the malfunction of one component could jeopardize the entire process, presents a challenge for unguided mechanisms. The mismatch repair system exemplified by the MutS, MutL, and MutH proteins in bacteria is another marvel. These proteins not only need to recognize mismatches that escape the proofreading capability of DNA polymerase but also have to signal for repair without causing unnecessary genomic instability. The accuracy and efficiency of this system are crucial for maintaining genetic fidelity. Double-strand breaks are perhaps one of the most daunting challenges for a cell. The emergence of repair mechanisms like homologous recombination and non-homologous end joining is essential. The coordination required between the various proteins involved, along with the need to choose the correct repair pathway based on the cellular context, is a complex task. The seamless operation of these systems in the LBCA stands in stark contrast to the potential absence or rudimentary nature of these mechanisms in LUCA. On the protection front, evolving sophisticated DNA-binding proteins that both protect and regulate DNA topology, like the bacterial histone-like proteins, is no minor feat. These proteins must interact with DNA in a manner that is both protective and functional, allowing for processes like transcription and replication to occur unhindered. Furthermore, the emergence of advanced antioxidant systems and enzymes to neutralize reactive oxygen species in the LBCA is essential. The specificity of these enzymes, along with their ability to neutralize a wide variety of reactive species without causing harm to the cell, presents a challenge to unguided evolutionary narratives.
Given the challenges associated with the evolution of sophisticated mechanisms from rudimentary prototypes, a polyphyletic creation proposition gains traction. The sheer complexity and interdependence of DNA repair and protection mechanisms present in the LBCA are difficult to account for through stepwise evolutionary processes. Each system is composed of multiple proteins and factors that must work in harmony. A slight deviation or malfunction in one component could jeopardize the entire system. The simultaneous emergence and fine-tuning of such components point towards a directed orchestration rather than a series of random, unguided events. The high specificity exhibited by enzymes involved in DNA repair, protection, and protein folding is remarkable. These entities are tailored to recognize, bind, and act upon very specific substrates or DNA lesions. Such precision, emerging from a backdrop of potential chemical chaos, seems more in line with a purposeful design rather than mere chance. Many of the discussed mechanisms exhibit a functional threshold, below which they might be non-operational or even detrimental to the cell. For systems that rely on multiple components, the stepwise addition of components over evolutionary time might render the intermediate stages non-functional or less beneficial. A polyphyletic creation or ID model posits that these systems were introduced in their functional entirety, bypassing the challenges associated with intermediate, potentially non-functional stages. The transition from LUCA to LBCA implies a significant increase in genetic information. The addition of new genes, regulatory elements, and metabolic pathways necessitates mechanisms that not only generate new genetic sequences but also ensure their integration and functionality within the existing genomic landscape. The directed input of information, as proposed by a creation or ID model, provides a more straightforward explanation for such increases. The DNA repair and protection mechanisms do not operate in isolation. They are interwoven with other cellular systems like replication, transcription, and translation. The harmonious integration of new and advanced mechanisms within an existing complex cellular milieu seems more congruent with a directed design. Given the complexities, intricacies, and interdependencies observed in the transition from LUCA to LBCA, a polyphyletic creation or Intelligent Design perspective presents a case-adequate, evidence-based alternative. This viewpoint posits that the emergence of advanced biological systems, exhibiting precision and functionality, might be better explained by directed processes or interventions rather than solely relying on unguided evolutionary mechanisms.
Other Proteins and Complexes
Venturing beyond the realm of familiar proteins in LUCA, one might find oneself enthralled by the vast ensemble of additional proteins and complexes that would potentially play their role in the LBCA. This world is akin to discovering an artist's expanded palette, each color nuanced, vibrant, and critical for painting the intricate tableau of bacterial life. In the primordial symphony of LUCA, some proteins might have had generalized roles, serving multiple functions like a multi-tool, adequate for the simpler cellular tasks of the time. Consider a carpenter's first toolkit – basic, functional, and vital. These might have been the proteins ensuring the rudimentary transport, signaling, and structural integrity of LUCA. However, as our narrative transitions to LBCA, the scenario becomes arguably more sophisticated. The proteins and complexes within the LBCA would have to be akin to an artist's specialized brushes or a master carpenter's refined tools. There would be proteins tailored for specific tasks, honed purportedly over countless generations to achieve maximal efficiency. The bacterial flagellum might stand out as an exemplary complex. While LUCA might not have possessed such intricate machinery, the LBCA would potentially flaunt a flagellum, a marvel of evolutionary engineering if it indeed evolved as such. This motor-driven apparatus would allow for directed movement, sensing gradients of nutrients or toxins in the environment. Envision a ship with a cutting-edge navigation system and a powerful engine, navigating the vast oceans with purpose and agility. In the same vein, bacterial pili, slender hair-like structures, might have become more specialized in LBCA, allowing for adherence, signaling, and even the exchange of genetic material. Think of these as the tendrils of a vine, each tendril optimized for a distinct function, anchoring the plant, sensing the environment, or reaching out to neighboring plants. Secretion systems would arguably be another marvel of LBCA, specialized protein complexes enabling the export of proteins or the injection of effectors into other cells. Imagine a series of gates, sluices, and catapults, each designed for precise delivery, ensuring that the cell interacts efficiently with its surroundings or potential prey. In this imagined journey from LUCA to LBCA, we'd be asked to appreciate the vast leap from a generalized toolkit to a collection of highly specialized instruments. This story would highlight the purported adaptive prowess of evolution, where life, in its bid to thrive, would have to evolve tools that aren’t just more numerous but exponentially more intricate. We'd be witnessing a tale of how a cell might not just adapt to its environment, but arguably master it, wielding a collection of protein tools and complexes with unparalleled artistry and precision.
In the framework of LUCA, proteins would have served generalized roles, akin to an artist using broad brush strokes to paint vast landscapes. These proteins are thought to have maintained rudimentary transport, signaling, and structural integrity. Transitioning to LBCA, the landscape of proteins and complexes would have required more detailed and specialized tools. Theoretically, instead of multifunctional proteins, a repertoire of specialized proteins would have emerged, tailored for distinct tasks. It is claimed that the LBCA would have sported a bacterial flagellum, representing a significant leap in evolutionary engineering. Such a structure would not just be for propulsion but would serve as a sensor, detecting gradients of nutrients or toxins. This development is likened to ships with state-of-the-art navigation systems navigating vast oceans with defined purpose. The bacterial pili are conceived as having undergone a specialization process by the time of the LBCA. These slender, hair-like structures are thought to have enabled adherence, signaling, and the transfer of genetic material. Conceptualized as tendrils of a vine, they would each serve a unique purpose - from anchoring the bacterium, sensing its surroundings, to interacting with neighboring cells. The LBCA's repertoire is suggested to have boasted advanced secretion systems, enabling precise export of proteins and injecting effectors into other cells. This would have been an intricate system, mirroring gates, sluices, and catapults, all tailored for ensuring effective cellular interactions with the environment. This hypothetical transition from LUCA to LBCA underscores a narrative of increasing complexity. The journey from a generalized toolkit in LUCA to a myriad of highly specialized instruments in LBCA demonstrates the narrative of evolution's potential to bring about sophisticated machinery. This would show how a cell might have evolved its arsenal, not just in quantity but in quality and precision, to adapt and possibly master its environment.
Given the vast leap from the generalized toolkit of LUCA to the specialized instruments of LBCA, the traditional model of gradual evolution becomes intensely scrutinized. Irreducibly complex systems, like the bacterial flagellum, would require multiple components to come together simultaneously to function. Such a simultaneous emergence challenges the conventional step-by-step evolutionary framework. Instead, a polyphyletic creation or ID event can more adequately explain the sudden appearance of such intricate machinery. The transition from LUCA to LBCA would demand the emergence of multiple novel and specialized protein complexes and systems. The bacterial pili's evolution, optimized for distinct functionalities such as adherence, signaling, or genetic material exchange, speaks of more than just adaptation. Such systems' emergence might seem improbable through slow, unguided processes alone. Polyphyletic creation or ID offers a perspective where these systems are introduced purposefully, fitting the specialized needs of a more advanced bacterial ancestor. The LBCA, as depicted, would exhibit advanced nucleotide recycling mechanisms and streamlined pathways, reflecting an optimized system operating at peak efficiency. Such rapid optimization, moving from LUCA's rudimentary processes to LBCA's refined mechanisms, might seem improbable in the conventional evolutionary timeline. Polyphyletic creation or ID can more fittingly account for the introduction of these optimized systems in a relatively condensed timeframe. In the speculated transition from LUCA to LBCA, systems don't just evolve in isolation. They would need to be interdependent and coordinated for the cell to function. The integration of multiple systems, each evolving in synchrony, would be a monumental challenge for traditional evolutionary explanations. A polyphyletic creation or ID viewpoint can provide a more satisfactory explanation for such synchronized emergence, positing that the systems were designed to function in tandem from their inception. The hypothetical transition from LUCA to LBCA would present with gaps, both in terms of missing intermediate forms and unexplained leaps in complexity. These gaps, difficult to address through conventional evolutionary mechanisms, become more understandable when viewed through the lens of polyphyletic creation or ID. This perspective can argue that the observed discontinuities are evidence of distinct creation or design events.