The RNA polymerase enzyme and its function, evidence of design
https://reasonandscience.catsboard.com/t2016-the-rna-polymerase-enzyme-and-its-function-evidence-of-design
DNA, the double-stranded molecule that carries genetic information and makes up chromosomes, transcribes when mRNA makes copy of a DNA strand. In this way, cells make proteins that help the cell do any number of things, they make proteins that are essential to the life of the organism. This process begins with an enzyme called RNA polymerase. It's this enzyme that splits the DNA strands in two: a template strand and a coding strand. RNA uses that coding strand to transcribe information in the DNA to make a variety of "gene products" supporting the organism-including more RNA polymerase. A complex system of molecules is doing very specific things in very specific places at very specific times at very specific rates.It's a complex arrangement of six chains of different amino acids—over a thousand per chain. If you change the structure or remove even a few of the amino acids, he says, the enzyme's function collapses.
To describe the genesis, divergence and complexity of life on Earth, one must understand two major transitions: 1) between the ancient RNA-protein world and LUCA (the last universal common ancestor); and 2) between LUCA and LECA (the last eukaryotic common ancestor). On Earth, life divides into three domains: bacteria, archaea and eukaryotes.
Information flow within living organisms requires genetic material in DNA to be read and deciphered. Because, within the double helix, DNA bases project inward, DNA is more stable than single-stranded RNA, in which bases are more exposed. LUCA, therefore, developed a more stable DNA genome than was possible in the RNA-protein world. The DNA double helix, however, is difficult to unzip, so proteins must act on DNA to separate strands. The relative security of a DNA genome compared to a fragmented RNA genome comes with conditions.
LUCA (~3.5 to 3.8 billion years ago) was one of the first organisms with a streamlined DNA genome. The Earth is only ~4.5 billion years old, so LUCA is very ancient and the time for the RNA-protein world that preceded LUCA was short. To read the genetic information within a DNA genome requires a mobile protein factory called RNA polymerase. To initiate RNA synthesis on a DNA template, RNA polymerases found in the three domains of life require protein factors that help them to recognize and open a promoter DNA sequence. In bacteria, sigma factors help RNA polymerases bind promoter DNA.
The evolutionary relationship, among bacterial sigma factors and archaeal TFB-TBP has not clearly been known.
Burton and Burton hypothesize that both sigma factors and TFB arose from a primordial initiation factor at LUCA with 4-HTH motifs, and bacteria and archaea diverged in evolution about the time of LUCA largely because of this difference in genome reading styles. Archaeal TFB lost 2-HTH motifs and gained compensating functions including cooperation with TBP. Because bacteria and archaea interpret (read; "transcribe") their DNA genomes differently to synthesize RNA, these organisms became distinct.
Question: how and why should that distinction have evolved ? the differences of transcriptions are hudge, eukaryotic cells use over 100 protein sub units.
Evolution of promoter DNA sequences has long been a mystery, but bacterial promoters somewhat resemble archaeal and eukaryotic promoters.
Its still today a mystery, and to the well intended its obvious why. The resemblance of one with the other means nothing.
very early in evolution, bacteria and archaea came to read and interpret genomic information in DNA in fundamentally different ways, causing bacteria and archaea to diverge into distinct domains.
Question: how did they " learn " that feat ??
At LECA (~1.6-2.5 billion years ago) an archaea fused with multiple bacteria to form the first eukaryote.
Cool story. How do they possibly know ? Answer : they don't. There are hudge differences between the two :
Gene expression is linked to RNA transcription, which cannot happen without RNA polymerase. However, this is where the similarities between prokaryote and eukaryote expression end.
Transcription in Bacteria
In bacteria, all transcription is performed by a single type of RNA polymerase. This polymerase contains four catalytic subunits and a single regulatory subunit known as sigma (s). Interestingly, several distinct sigma factors have been identified, and each of these oversees transcription of a unique set of genes. Sigma factors are thus discriminatory, as each binds a distinct set of promoter sequences.
Transcription in Eukaryotes
Eukaryotic cells are more complex than bacteria in many ways, including in terms of transcription.
Specifically, in eukaryotes, transcription is achieved by three different types of RNA polymerase
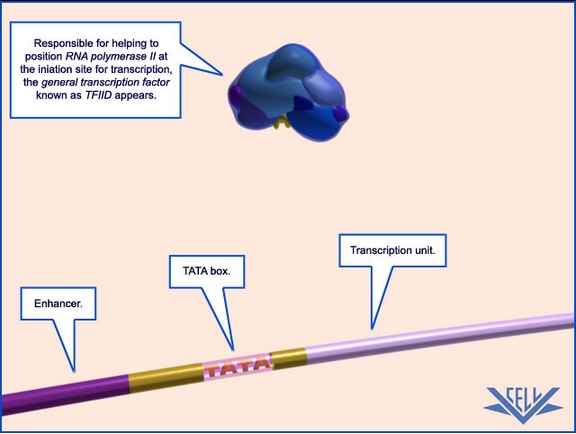
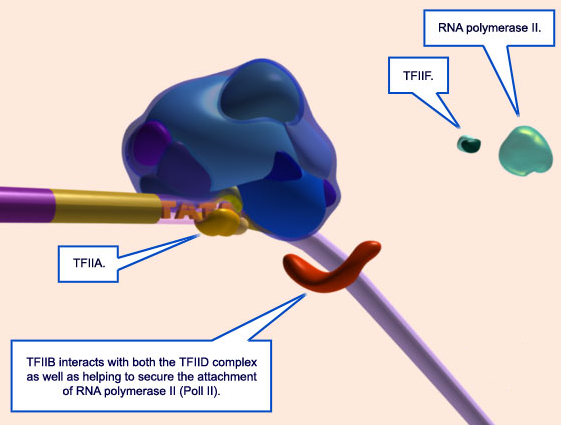
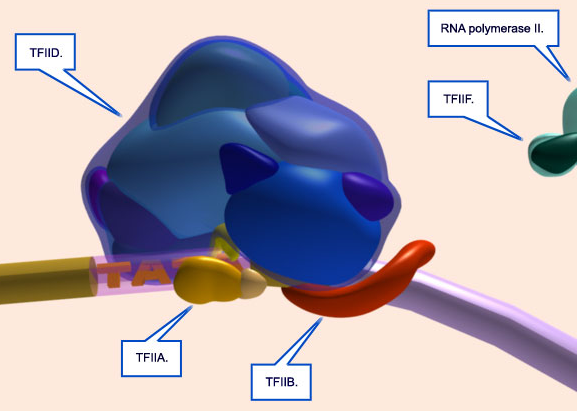
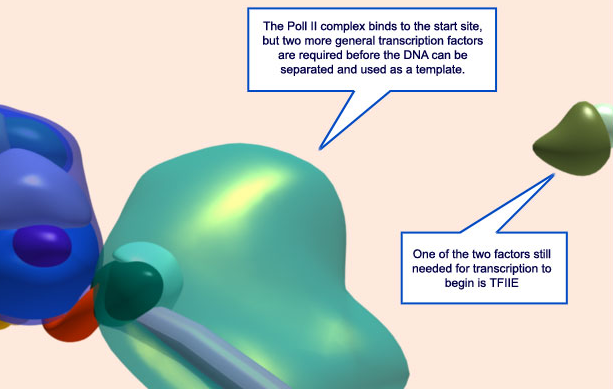
These polymerases differ in the number and type of subunits they contain, as well as the class of RNAs they transcribe; that is, RNA pol I transcribes ribosomal RNAs (rRNAs), RNA pol II transcribes RNAs that will become messenger RNAs (mRNAs) and also small regulatory RNAs, and RNA pol III transcribes small RNAs such as transfer RNAs (tRNAs). Because RNA pol II transcribes protein-encoding genes, it has been of particular importance to scientists who study the regulation of eukaryotic gene expression, and its function is well understood. For example, researchers know that RNA pol II can bind to a DNA sequence within the promoter of many genes, known as the TATA box, to initiate transcription.
So there are remarkable differences and a dramatic increase of complexity of this machinery between prokaryotic and eukaryotic cells. Its hard to see how and why there could have been the evolution from one to the other 3 in eukaryotic cells.
Together with other common motifs (short recognition sequences in the DNA), these elements constitute the core promoter.
Bacterial RNA polymerase is a huge factory with many moving parts. It is composed of a dozen different proteins. Together, they form a machine that surrounds DNA strands, unwinds them, and builds an RNA strand based on the information held inside the DNA. Once the enzyme gets started, RNA polymerase marches confidently along the DNA copying RNA strands thousands of nucleotides long.
Observe the nomenclature : huge factory, machine, marches. Question : how did the dozens of proteins arise ? Unless this protein machine was not fully formed, no mRNA could be formed, no transcription and subsequently, no replication would be possible, upon which mutations and natural selection could act. So the only alternatives left are chance, physical necessity, and design. Could it be that chance, or physical necessity are good explanations for the arise of the enormously complex RNA polymerase machine?
Accuracy
As you might expect, RNA polymerase needs to be accurate in its copying of genetic information. To improve its accuracy, it performs a simple proofreading stepas it builds an RNA strand.
Question: unless enough transcription accuracy is achieved, the whole process would not be accurate enough, and the whole process would be compromised. How could it have got this amazing feature upon natural ways ? Isnt that far better explained through a intentional designer, who created this essential mechanism ?
The active site is designed to be able to remove nucleotides as well as add them to the growing strand. The enzyme tends to hover around mismatched nucleotides longer than properly added ones, giving the enzyme time to remove them.
Question : how did the machine arise this amazing ability ?
This process is somewhat wasteful, since proper nucleotides are also occasionally removed, but this is a small price to pay for creating better RNA transcripts. Overall, RNA polymerase makes an error about once in 10,000 nucleotides added, or about once per RNA strand created.
Question: How did it get such a amazing accuracy ?
To transcribe a gene accurately, RNA polymerase must recognize where on the genome to start and where to finish.
Question: Had RNA polymerase not to be able to recognize where to start right from the beginning, otherwise it would not be functional ? How did it learn that feat ?
The initiation of transcription is an especially important step in gene expression because it is the main point at which the cell regulates which proteins are to be produced and at what rate.
Question : had the cell not to be able right from the start of its operation to regulate which proteins had to be produced, and and what rate ? How did it " learn " that feat ? Unless it was able to do so right from the beginning, would chaos not be the result ? Would the cell not cease to function properly ?
The bacterial RNA polymerase core enzyme is a multisubunit complex that sythesizes RNA using a DNA template as a guide.
Question : Unless it uses a DNA template as a guide, would the whole process also not be functional ? Had it not have to be able to use the DNA template as a guide right from the start ?
A detachable subunit called sigma (s) factor associates with the core enzyme and assists it in reading the signals in the DNA that tell it where to begin transcribingThese proteins belong to a family of polypeptides and they have very interesting features and play important role in specific gene expression.
They associate with RNAP subunits and they are also responsible for identifying the correct promoter and positioning the RNAP on DNA in proper context for initiating transcription. Otherwise the RNAP without sig cannot function as an enzyme; it is like a driver without a rudder/conductor.
Sigma (s) is a dissociable subunit of RNAP. When s is bound to core, the resulting complex, called holoenzyme, can bind with high affinity to promoter sites.
Without the sigma factor, RNA polymerase would bind loosely and randomly to DNA, but would not be able to start transcription. Therefore no proteins would be made and the cell would die. 4)
Question: the sigma factor alone is LIFE ESSENTIAL. Had it not have to be there fully functional and programmed to do its job right when life started ?
It seems the RNA polymerase enzyme could not be a product of chance,
necessity (or natural law), or a combination of chance and necessity, and neither evolution, since evolution only works after replication is in place. The best probabilistic explanation is therefore design. Is this a argument from ignorance ? I don't think so.
http://www.arn.org/docs/booher/scientific-case-for-ID.html
This objection is essentially stating that intelligent design is just a mysterious notion that covers for ignorance—a sort of God of the gaps argument. But the design inference does not constitute an argument from ignorance, rather it constitutes an “inference to the best explanation” based on our best available knowledge. “We are not ignorant of how information arises. We know from experience that conscious intelligent agents can create informational sequences and systems…. When we encounter the information based in large biological molecules needed for life, we can infer—based on our knowledge of established cause and effect relationship—that an intelligent cause operated in the past to produce the specified information necessary to the origin of life.” Intelligent design is applying a standard historical sciences principle of uniformitarianism, that “the present is the key to the past.”
DNA, the double-stranded molecule that carries genetic information and makes up chromosomes, transcribes when mRNA makes copy of a DNA strand. In this way, cells make proteins that help the cell do any number of things, they make proteins that are essential to the life of the organism. This process begins with an enzyme called RNA polymerase. It's this enzyme that splits the DNA strands in two: a template strand and a coding strand. RNA uses that coding strand to transcribe information in the DNA to make a variety of "gene products" supporting the organism-including more RNA polymerase. A complex system of molecules is doing very specific things in very specific places at very specific times at very specific rates.It's a complex arrangement of six chains of different amino acids—over a thousand per chain. If you change the structure or remove even a few of the amino acids, he says, the enzyme's function collapses 11
To describe the genesis, divergence and complexity of life on Earth, one must understand two major transitions: 1) between the ancient RNA-protein world and LUCA (the last universal common ancestor); and 2) between LUCA and LECA (the last eukaryotic common ancestor). On Earth, life divides into three domains: bacteria, archaea and eukaryotes. 7
Information flow within living organisms requires genetic material in DNA to be read and deciphered. Because, within the double helix, DNA bases project inward, DNA is more stable than single-stranded RNA, in which bases are more exposed. LUCA, therefore, developed a more stable DNA genome than was possible in the RNA-protein world. The DNA double helix, however, is difficult to unzip, so proteins must act on DNA to separate strands. The relative security of a DNA genome compared to a fragmented RNA genome comes with conditions.
LUCA (~3.5 to 3.8 billion years ago) was one of the first organisms with a streamlined DNA genome. The Earth is only ~4.5 billion years old, so LUCA is very ancient and the time for the RNA-protein world that preceded LUCA was short. To read the genetic information within a DNA genome requires a mobile protein factory called RNA polymerase. To initiate RNA synthesis on a DNA template, RNA polymerases found in the three domains of life require protein factors that help them to recognize and open a promoter DNA sequence. In bacteria, sigma factors help RNA polymerases bind promoter DNA.
The evolutionary relationship, among bacterial sigma factors and archaeal TFB-TBP has not clearly been known.
Burton and Burton hypothesize that both sigma factors and TFB arose from a primordial initiation factor at LUCA with 4-HTH motifs, and bacteria and archaea diverged in evolution about the time of LUCA largely because of this difference in genome reading styles. Archaeal TFB lost 2-HTH motifs and gained compensating functions including cooperation with TBP. Because bacteria and archaea interpret (read; "transcribe") their DNA genomes differently to synthesize RNA, these organisms became distinct.
Question: how and why should that distinction have evolved ? the differences of transcriptions are hudge, eukaryotic cells use over 100 protein sub units.
Evolution of promoter DNA sequences has long been a mystery, but bacterial promoters somewhat resemble archaeal and eukaryotic promoters.
Its still today a mystery, and to the well intended its obvious why. The resemblance of one with the other means nothing.
very early in evolution, bacteria and archaea came to read and interpret genomic information in DNA in fundamentally different ways, causing bacteria and archaea to diverge into distinct domains.
Question: how did they " learn " that feat ??
At LECA (~1.6-2.5 billion years ago) an archaea fused with multiple bacteria to form the first eukaryote.
Cool story. How do they possibly know ? Answer : they don't. There are hudge differences between the two :
Gene expression is linked to RNA transcription, which cannot happen without RNA polymerase. However, this is where the similarities between prokaryote and eukaryote expression end. 8
Transcription in Bacteria
In bacteria, all transcription is performed by a single type of RNA polymerase. This polymerase contains four catalytic subunits and a single regulatory subunit known as sigma (s). Interestingly, several distinct sigma factors have been identified, and each of these oversees transcription of a unique set of genes. Sigma factors are thus discriminatory, as each binds a distinct set of promoter sequences.
Transcription in Eukaryotes
Eukaryotic cells are more complex than bacteria in many ways, including in terms of transcription.
Specifically, in eukaryotes, transcription is achieved by three different types of RNA polymerase
These polymerases differ in the number and type of subunits they contain, as well as the class of RNAs they transcribe; that is, RNA pol I transcribes ribosomal RNAs (rRNAs), RNA pol II transcribes RNAs that will become messenger RNAs (mRNAs) and also small regulatory RNAs, and RNA pol III transcribes small RNAs such as transfer RNAs (tRNAs). Because RNA pol II transcribes protein-encoding genes, it has been of particular importance to scientists who study the regulation of eukaryotic gene expression, and its function is well understood. For example, researchers know that RNA pol II can bind to a DNA sequence within the promoter of many genes, known as the TATA box, to initiate transcription.
So there are remarkable differences and a dramatic increase of complexity of this machinery between prokaryotic and eukaryotic cells. Its hard to see how and why there could have been the evolution from one to the other 3 in eukaryotic cells.
Together with other common motifs (short recognition sequences in the DNA), these elements constitute the core promoter.
So there is not only a difference between the RNA polymerase machinery, but also in the promoters sections in DNA.
Prokaryotic vs. Eukaryotic Trancription
Differences:
Prokaryotes only contain three different promoter elements: -10, -35 promoters, and upstream elements. Eukaryotes contain many different promoter elements: TATA box, initiator elements, downstream core promoter element, CAAT box, and the GC box to name a few. Eukaryotes have three types of RNA polymerases, I, II, and III, and prokaryotes only have one type. Eukaryotes form and initiation complex with the various transcription factors that dissociate after initiation is completed. There is no such structure seen in prokaryotes. Another main difference between the two is that transcription and translation occurs simultaneously in prokaryotes and in eukaryotes the RNA is first transcribed in the nucleus and then translated in the cytoplasm. RNAs from eukaryotes undergo post-transcriptional modifications including: capping, polyadenylation, and splicing. These events do not occur in prokaryotes. mRNAs in prokaryotes tend to contain many different genes on a single mRNA meaning they are polycystronic. Eukaryotes contain mRNAs that are monocystronic. Termination in prokaryotes is done by either rho-dependent or rho-independent mechanisms. In eukaryotes transcription is terminated by two elements: a poly(A) signal and a downstream terminator sequence. 9
These are hudge differences. How could or should the transition from one kind of promoters and RNA polymerase machinery have occured to the other domain, far more complex ?
The RNA in a cell is made by DNA transcription, a process that has certain similarities to the process of DNA replication. The machines doing this job, RNA polymerase enzymes, are essential to life and are found in all organisms and many viruses.
By selecting which RNAs are made, RNA polymerase (RNAP) dictates how cells adapt to new environments, interact symbiotically or pathogenically with hosts, respond to stress and starvation, and multiply. RNAP accomplishes this task by deciding where and how often to start transcription, how to elongate RNAs, and where to stop. 6
Question: How did the cell " learn " how to decide where, and how often to start transcription ? Had this mechanism not have to be in place before the first cell was ready to replicate ?
Emergence of transcription initiation factors in the three domains of life.
The bacterial RNA polymerase (RNAP) strictly depends on σ-factors for transcription initiation, whereas archaeal and eukaryotic RNAPs require the initiation factors TATA box-binding protein (TBP) and transcription factor IIB (TFIIB). There are four potential scenarios for the evolution of these factors. As the first scenario requires the fewest independent evolutionary events, it is the simplest explanation for the occurrence of transcription initiation factors in all extant life. a | As there are no σ-factor homologues in archaea and eukaryotes, and no TBP and TFIIB homologues in bacteria, it is likely that the ancestral RNAP of the last universal common ancestor (LUCA) used neither σ-factors nor TBP or TFIIB factors, and that these factors evolved independently in the bacterial and archaeal–eukaryotic lineages, respectively, after their split. b | An alternative scenario is that the RNAP of the LUCA used both σ-factors and TBP and TFIIB factors in parallel, and then lost the relevant factors in each lineage. c | A third scenario is that the LUCA used σ-factors, and then the archaeal–eukaryotic lineage lost these factors are gained TBP and TFIIB factors. d | The final scenario is that the LUCA used TBP and TFIIB factors, and that these were then lost in the bacterial lineage and σ-factors were gained.10

There are four potential scenarios for the evolution of these transcription factors. Translated : they have no idea how the different factors could have evolved , and the mechanisms involved in a detailled manner. They just make things up. If someone expected to read the paper on how this large, complex transcription machine evolved, you would be disappointed. The authors merely assume it evolved, and attempt to place various scenarios into an evolutionary tree where they don’t fit very well. This is typical of evolutionary scientific literature that already “knows” evolution is a “fact” but has no way to back up the claim. Highly-complex, interdependent systems that involves a coded , specified, complex language, translation, regulation, feedback, quality control and highest precision. The paper above fails completely to address the origin of the most efficient information-processing system known on earth. And so do all mainstream scientific papers. Their frequent answer is : " We don't know yet ". As if one day they would know, and evolution would be confirmed...... Wait a minute. Really ??
Bacterial RNA polymerase is a huge factory with many moving parts. It is composed of a dozen different proteins. Together, they form a machine that surrounds DNA strands, unwinds them, and builds an RNA strand based on the information held inside the DNA. Once the enzyme gets started, RNA polymerase marches confidently along the DNA copying RNA strands thousands of nucleotides long. 1
Observe the nomenclature : huge factory, machine, marches. Question : how did the dozens of proteins arise ? Unless this protein machine was not fully formed, no mRNA could be formed, no transcription and subsequently, no replication would be possible, upon which mutations and natural selection could act. So the only alternatives left are chance, physical necessity, and design. Could it be that chance, or physical necessity are good explanations for the arise of the enormously complex RNA polymerase machine?
Accuracy
As you might expect, RNA polymerase needs to be accurate in its copying of genetic information. To improve its accuracy, it performs a simple proofreading stepas it builds an RNA strand.
Question: unless enough transcription accuracy is achieved, the whole process would not be accurate enough, and the whole process would be compromised. How could it have got this amazing feature upon natural ways ? Isnt that far better explained through a intentional designer, who created this essential mechanism ?
The active site is designed to be able to remove nucleotides as well as add them to the growing strand. The enzyme tends to hover around mismatched nucleotides longer than properly added ones, giving the enzyme time to remove them.
Question : how did the machine arise this amazing ability ?
This process is somewhat wasteful, since proper nucleotides are also occasionally removed, but this is a small price to pay for creating better RNA transcripts. Overall, RNA polymerase makes an error about once in 10,000 nucleotides added, or about once per RNA strand created.
Question: How did it get such a amazing accuracy ?
To transcribe a gene accurately, RNA polymerase must recognize where on the genome to start and where to finish.
Question: Had RNA polymerase not to be able to recognize where to start right from the beginning, otherwise it would not be functional ? How did it learn that feat ?
The initiation of transcription is an especially important step in gene expression because it is the main point at which the cell regulates which proteins are to be produced and at what rate.
Question : had the cell not to be able right from the start of its operation to regulate which proteins had to be produced, and and what rate ? How did it " learn " that feat ? Unless it was able to do so right from the beginning, would chaos not be the result ? Would the cell not cease to function properly ?
The bacterial RNA polymerase core enzyme is a multisubunit complex that sythesizes RNA using a DNA template as a guide.
Question : Unless it uses a DNA template as a guide, would the whole process also not be functional ? Had it not have to be able to use the DNA template as a guide right from the start ?
A detachable subunit called sigma (s) factor associates with the core enzyme and assists it in reading the signals in the DNA that tell it where to begin transcribing
Sigma factors:
http://en.wikipedia.org/wiki/Sigma_factor
These proteins belong to a family of polypeptides and they have very interesting features and play important role in specific gene expression.
They associate with RNAP subunits and they are also responsible for identifying the correct promoter and positioning the RNAP on DNA in proper context for initiating transcription. Otherwise the RNAP without sig cannot function as an enzyme; it is like a driver without a rudder/conductor.
Sigma (s) is a dissociable subunit of RNAP. When s is bound to core, the resulting complex, called holoenzyme, can bind with high affinity to promoter sites. 3)
Without the sigma factor, RNA polymerase would bind loosely and randomly to DNA, but would not be able to start transcription. Therefore no proteins would be made and the cell would die. 4)
Question: the sigma factor alone is LIFE ESSENTIAL. Had it not have to be there fully functional and programmed to do its job right when life started ?
Core RNA polymerase in bacteria is a complex composed of an
two alpha subunits
one beta, one beta’
one omega subunit,
together they form the core structure.
The α-amino-terminal domains (α-NTDs) serve as a scaffold for complex assembly;
the α-carboxy-terminal domains (α-CTDs) and ω-subunit play regulatory roles during initiation.
The β- and β′-subunits jointly form the active site and make all the contacts to the nucleic acids.
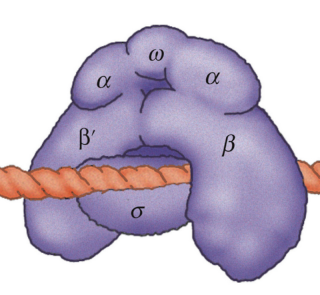
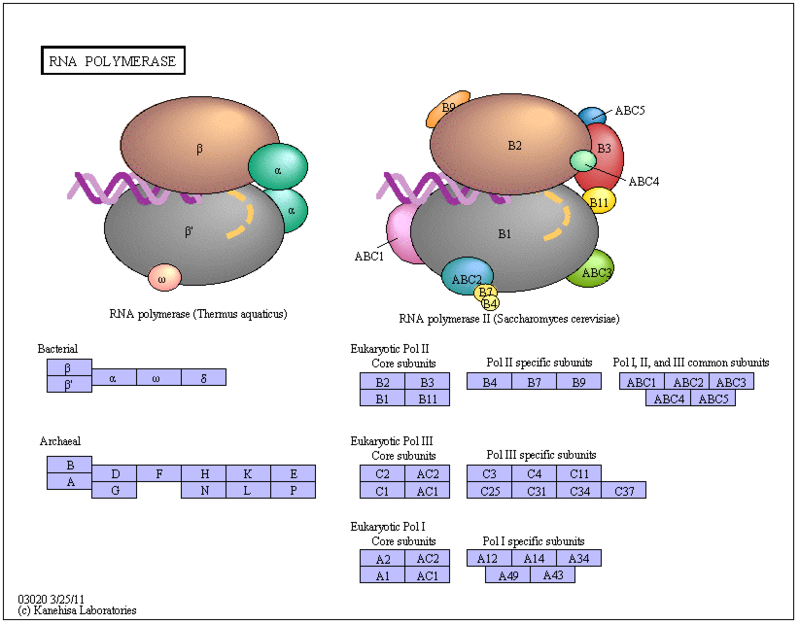
image ru
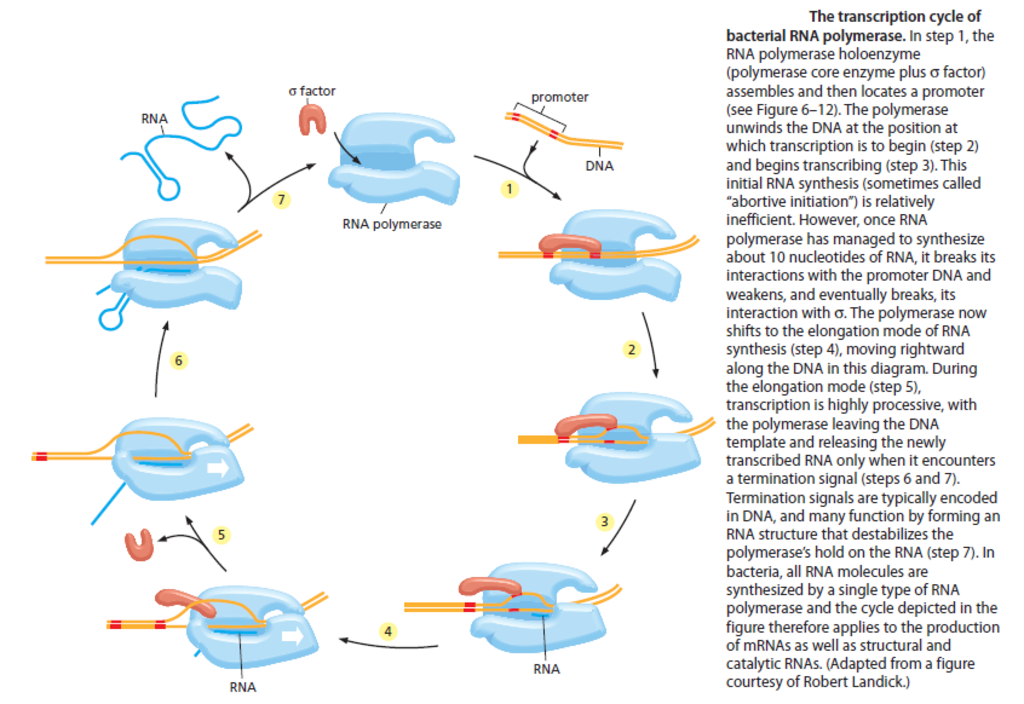
The process of transcription can be divided into three stages:
1) Initiation of a new RNA chain (parts 1-4 of the picture below).
2) Elongation of the chain (parts 5&6 of the picture below).
3) Termination of transcription and release of the brand new RNA molecule (parts 6&7 of the picture below).
upload pic
Together, s factor and core enzyme are known as the RNA polymerase holoenzyme; this complex adheres only weakly to bacterial DNA when the two collide, and a holoenzyme typically slides rapidly along the long DNA molecule until it dissociates again. However, when the polymerase holoenzyme slides into a region on the DNA double helix called a promoter, a special sequence of nucleotides indicating the starting point for RNA synthesis, the polymerase binds tightly to this DNA.
Question: Is it not essential, that this special sequence of nucleotides is there in the DNA code right from the beginning, otherwise the right mRNA cannot be synthesized ? How did these special sequences get there naturally ? How did the polymerase " learn " to bind tightly to this DNA ? Had it not have to be able to bind tightly right from the beginning ? Are these not functions, that are essential, and if they are not fully functional right from the beginning, they do not function at all ?
The polymerase holoenzyme, through its s factor, recognizes the promoter DNA sequence by making specific contacts with the portions of the bases that are exposed on the outside of the helix step 1 in the Figure above. After the RNA polymerase holoenzyme binds tightly to the promoter DNA in this way, it opens up the double helix to expose a short stretch of nucleotides on each strand (step 2 )
Question: how did the holoenzyme " learn " to do this precise step in a functional and correct way ? Try and fail attempts would not permit the process to go ahead, and the whole process would be compromised. Had the holoenzyme not have to be functional and fully formed right from the beginning ?
With the DNA unwound, one of the two exposed DNA strands acts as a template for complementary base-pairing with incoming ribonucleotides, two of which are joined together by the polymerase to begin an RNA chain (step 3 )

Questions:
Had the incoming and exit movement of the DNA strand not have to be pre programmed ?
Had the enter and exit channel of DNA not have to be precisely designed, and be there right from the beginning ? There could be no function of a incompletely designed enzyme.
Had the incoming and exit movement of the ribonucleotides and newly formed mRNA strand not have to be pre programmed ?
Had the enter and exit channel of ribonucleotides and the mRNA strand not have to be precisely designed, and be there right from the beginning ?
Had the ribonucleotides not have to be readily availabe for recruitment right from the beginning ? Had the entry channel and the exit channel of the ribonucleotides to enter and leave the factory and being moved and released to the place where the action goes on not to be designed?
After the first ten or so nucleotides of RNA have been synthesized (a relatively inefficient process during which polymerase synthesizes and discards short RNA oligomers), the core enzyme breaks its interactions with the promoter DNA, weakens its interactions with s factor, and begins to move down the DNA, synthesizing RNA (steps 4 and 5 )
Chain elongation continues (at a speed of approximately 50 nucleotides/sec for bacterial RNA polymerases) until the enzyme encounters a second signal in the DNA, the terminator , where the polymerase halts and releases both the newly made RNA chain and the DNA template (step 7 ) After the polymerase core enzyme has been released at a terminator, it reassociates with a free s factor to form a holoenzyme that can begin the process of transcription again.
Termination signals induce rapid and irreversible dissociation of the nascent transcript from RNA polymerase. Terminators at the end of genes prevent unintended transcription into the downstream genes, whereas terminators in the upstream regulatory leader regions adjust expression of the structural genes in response to metabolic and environmental signals. Premature termination within an operon leads to potentially deleterious defects in the expression of the downstream genes, but also provides an important surveillance mechanism. 5
Question : had the terminator not have to be there right from the beginning, otherwise the new mRNA strand would not have the right end ?
The processes of transcription initiation and termination involve a complicated series of structural transitions in protein, DNA, and RNA molecules. Unless the the whole operational sequence of the transcription cycle is not fully functional right from the start, transcription cannot occur. Take away just one of the steps, and the process ceases to function. Is it feasable and imaginable, that the RNA polymerase enzyme with its complex transcription process requiring many parts and subunits could be the product of random natural processes ? Transcription is a irreducible process. Furthermore, without replication , there is no evolution. In order for replication to occur, the DNA polymerase and RNA polymerase machinery had to be fully operational right since the start of life, so the machinery had to exist prior mutations and natural selection could occur.
Core promotors have to be encoded in DNA, otherwise the RNA polymerase enzyme does not know where to start and end transcription. Without it, no life would exist. It is impossible that it got there by trial and error. Unless it was at the right place right from the start, life could not happen. Feel free to bring chance into the game or good luck. You are guillable to the extreme, doing so. Either its fully functional, or it does not function at all. Is that not strong evidence for intelligent design ? If not, what explanation of origin fits better the observation described here ? [/justify]
http://reasonandscience.heavenforum.org/t2016-the-rna-polymerase-enzyme-and-its-function-evidence-of-design#3391
1) http://www.rcsb.org/pdb/101/motm.do?momID=40
2) http://www.proteopedia.org/wiki/index.php/2e2i
3) http://www.researchgate.net/profile/John_Helmann/publication/228031784_Sigma_Factors_in_Gene_Expression/links/0fcfd506ac99963842000000.pdf
4) http://helicase.pbworks.com/w/page/17605688/Matthew-Hunter
5) http://www.nature.com/nrmicro/journal/v9/n5/abs/nrmicro2560.html
6) http://landick.wisc.edu/images/geszvain_landick.pdf
7) https://bmb.natsci.msu.edu/about/directory/faculty/zachary-f-burton/genesis-of-life-on-earth/
8 )http://www.nature.com/scitable/topicpage/rna-transcription-by-rna-polymerase-prokaryotes-vs-961
9) http://www.chem.uwec.edu/webpapers2006/sites/demlba/folder/provseuk.html
10) http://www.nature.com/nrmicro/journal/v9/n2/fig_tab/nrmicro2507_F8.html
11) http://storage.cloversites.com/makinglifecountministriesinc/documents/Intelligent%20Design.pdf
https://reasonandscience.catsboard.com/t2016-the-rna-polymerase-enzyme-and-its-function-evidence-of-design
DNA, the double-stranded molecule that carries genetic information and makes up chromosomes, transcribes when mRNA makes copy of a DNA strand. In this way, cells make proteins that help the cell do any number of things, they make proteins that are essential to the life of the organism. This process begins with an enzyme called RNA polymerase. It's this enzyme that splits the DNA strands in two: a template strand and a coding strand. RNA uses that coding strand to transcribe information in the DNA to make a variety of "gene products" supporting the organism-including more RNA polymerase. A complex system of molecules is doing very specific things in very specific places at very specific times at very specific rates.It's a complex arrangement of six chains of different amino acids—over a thousand per chain. If you change the structure or remove even a few of the amino acids, he says, the enzyme's function collapses.
To describe the genesis, divergence and complexity of life on Earth, one must understand two major transitions: 1) between the ancient RNA-protein world and LUCA (the last universal common ancestor); and 2) between LUCA and LECA (the last eukaryotic common ancestor). On Earth, life divides into three domains: bacteria, archaea and eukaryotes.
Information flow within living organisms requires genetic material in DNA to be read and deciphered. Because, within the double helix, DNA bases project inward, DNA is more stable than single-stranded RNA, in which bases are more exposed. LUCA, therefore, developed a more stable DNA genome than was possible in the RNA-protein world. The DNA double helix, however, is difficult to unzip, so proteins must act on DNA to separate strands. The relative security of a DNA genome compared to a fragmented RNA genome comes with conditions.
LUCA (~3.5 to 3.8 billion years ago) was one of the first organisms with a streamlined DNA genome. The Earth is only ~4.5 billion years old, so LUCA is very ancient and the time for the RNA-protein world that preceded LUCA was short. To read the genetic information within a DNA genome requires a mobile protein factory called RNA polymerase. To initiate RNA synthesis on a DNA template, RNA polymerases found in the three domains of life require protein factors that help them to recognize and open a promoter DNA sequence. In bacteria, sigma factors help RNA polymerases bind promoter DNA.
The evolutionary relationship, among bacterial sigma factors and archaeal TFB-TBP has not clearly been known.
Burton and Burton hypothesize that both sigma factors and TFB arose from a primordial initiation factor at LUCA with 4-HTH motifs, and bacteria and archaea diverged in evolution about the time of LUCA largely because of this difference in genome reading styles. Archaeal TFB lost 2-HTH motifs and gained compensating functions including cooperation with TBP. Because bacteria and archaea interpret (read; "transcribe") their DNA genomes differently to synthesize RNA, these organisms became distinct.
Question: how and why should that distinction have evolved ? the differences of transcriptions are hudge, eukaryotic cells use over 100 protein sub units.
Evolution of promoter DNA sequences has long been a mystery, but bacterial promoters somewhat resemble archaeal and eukaryotic promoters.
Its still today a mystery, and to the well intended its obvious why. The resemblance of one with the other means nothing.
very early in evolution, bacteria and archaea came to read and interpret genomic information in DNA in fundamentally different ways, causing bacteria and archaea to diverge into distinct domains.
Question: how did they " learn " that feat ??
At LECA (~1.6-2.5 billion years ago) an archaea fused with multiple bacteria to form the first eukaryote.
Cool story. How do they possibly know ? Answer : they don't. There are hudge differences between the two :
Gene expression is linked to RNA transcription, which cannot happen without RNA polymerase. However, this is where the similarities between prokaryote and eukaryote expression end.
Transcription in Bacteria
In bacteria, all transcription is performed by a single type of RNA polymerase. This polymerase contains four catalytic subunits and a single regulatory subunit known as sigma (s). Interestingly, several distinct sigma factors have been identified, and each of these oversees transcription of a unique set of genes. Sigma factors are thus discriminatory, as each binds a distinct set of promoter sequences.
Transcription in Eukaryotes
Eukaryotic cells are more complex than bacteria in many ways, including in terms of transcription.
Specifically, in eukaryotes, transcription is achieved by three different types of RNA polymerase
These polymerases differ in the number and type of subunits they contain, as well as the class of RNAs they transcribe; that is, RNA pol I transcribes ribosomal RNAs (rRNAs), RNA pol II transcribes RNAs that will become messenger RNAs (mRNAs) and also small regulatory RNAs, and RNA pol III transcribes small RNAs such as transfer RNAs (tRNAs). Because RNA pol II transcribes protein-encoding genes, it has been of particular importance to scientists who study the regulation of eukaryotic gene expression, and its function is well understood. For example, researchers know that RNA pol II can bind to a DNA sequence within the promoter of many genes, known as the TATA box, to initiate transcription.
So there are remarkable differences and a dramatic increase of complexity of this machinery between prokaryotic and eukaryotic cells. Its hard to see how and why there could have been the evolution from one to the other 3 in eukaryotic cells.
Together with other common motifs (short recognition sequences in the DNA), these elements constitute the core promoter.
Bacterial RNA polymerase is a huge factory with many moving parts. It is composed of a dozen different proteins. Together, they form a machine that surrounds DNA strands, unwinds them, and builds an RNA strand based on the information held inside the DNA. Once the enzyme gets started, RNA polymerase marches confidently along the DNA copying RNA strands thousands of nucleotides long.
Observe the nomenclature : huge factory, machine, marches. Question : how did the dozens of proteins arise ? Unless this protein machine was not fully formed, no mRNA could be formed, no transcription and subsequently, no replication would be possible, upon which mutations and natural selection could act. So the only alternatives left are chance, physical necessity, and design. Could it be that chance, or physical necessity are good explanations for the arise of the enormously complex RNA polymerase machine?
Accuracy
As you might expect, RNA polymerase needs to be accurate in its copying of genetic information. To improve its accuracy, it performs a simple proofreading stepas it builds an RNA strand.
Question: unless enough transcription accuracy is achieved, the whole process would not be accurate enough, and the whole process would be compromised. How could it have got this amazing feature upon natural ways ? Isnt that far better explained through a intentional designer, who created this essential mechanism ?
The active site is designed to be able to remove nucleotides as well as add them to the growing strand. The enzyme tends to hover around mismatched nucleotides longer than properly added ones, giving the enzyme time to remove them.
Question : how did the machine arise this amazing ability ?
This process is somewhat wasteful, since proper nucleotides are also occasionally removed, but this is a small price to pay for creating better RNA transcripts. Overall, RNA polymerase makes an error about once in 10,000 nucleotides added, or about once per RNA strand created.
Question: How did it get such a amazing accuracy ?
To transcribe a gene accurately, RNA polymerase must recognize where on the genome to start and where to finish.
Question: Had RNA polymerase not to be able to recognize where to start right from the beginning, otherwise it would not be functional ? How did it learn that feat ?
The initiation of transcription is an especially important step in gene expression because it is the main point at which the cell regulates which proteins are to be produced and at what rate.
Question : had the cell not to be able right from the start of its operation to regulate which proteins had to be produced, and and what rate ? How did it " learn " that feat ? Unless it was able to do so right from the beginning, would chaos not be the result ? Would the cell not cease to function properly ?
The bacterial RNA polymerase core enzyme is a multisubunit complex that sythesizes RNA using a DNA template as a guide.
Question : Unless it uses a DNA template as a guide, would the whole process also not be functional ? Had it not have to be able to use the DNA template as a guide right from the start ?
A detachable subunit called sigma (s) factor associates with the core enzyme and assists it in reading the signals in the DNA that tell it where to begin transcribingThese proteins belong to a family of polypeptides and they have very interesting features and play important role in specific gene expression.
They associate with RNAP subunits and they are also responsible for identifying the correct promoter and positioning the RNAP on DNA in proper context for initiating transcription. Otherwise the RNAP without sig cannot function as an enzyme; it is like a driver without a rudder/conductor.
Sigma (s) is a dissociable subunit of RNAP. When s is bound to core, the resulting complex, called holoenzyme, can bind with high affinity to promoter sites.
Without the sigma factor, RNA polymerase would bind loosely and randomly to DNA, but would not be able to start transcription. Therefore no proteins would be made and the cell would die. 4)
Question: the sigma factor alone is LIFE ESSENTIAL. Had it not have to be there fully functional and programmed to do its job right when life started ?
It seems the RNA polymerase enzyme could not be a product of chance,
necessity (or natural law), or a combination of chance and necessity, and neither evolution, since evolution only works after replication is in place. The best probabilistic explanation is therefore design. Is this a argument from ignorance ? I don't think so.
http://www.arn.org/docs/booher/scientific-case-for-ID.html
This objection is essentially stating that intelligent design is just a mysterious notion that covers for ignorance—a sort of God of the gaps argument. But the design inference does not constitute an argument from ignorance, rather it constitutes an “inference to the best explanation” based on our best available knowledge. “We are not ignorant of how information arises. We know from experience that conscious intelligent agents can create informational sequences and systems…. When we encounter the information based in large biological molecules needed for life, we can infer—based on our knowledge of established cause and effect relationship—that an intelligent cause operated in the past to produce the specified information necessary to the origin of life.” Intelligent design is applying a standard historical sciences principle of uniformitarianism, that “the present is the key to the past.”
DNA, the double-stranded molecule that carries genetic information and makes up chromosomes, transcribes when mRNA makes copy of a DNA strand. In this way, cells make proteins that help the cell do any number of things, they make proteins that are essential to the life of the organism. This process begins with an enzyme called RNA polymerase. It's this enzyme that splits the DNA strands in two: a template strand and a coding strand. RNA uses that coding strand to transcribe information in the DNA to make a variety of "gene products" supporting the organism-including more RNA polymerase. A complex system of molecules is doing very specific things in very specific places at very specific times at very specific rates.It's a complex arrangement of six chains of different amino acids—over a thousand per chain. If you change the structure or remove even a few of the amino acids, he says, the enzyme's function collapses 11
To describe the genesis, divergence and complexity of life on Earth, one must understand two major transitions: 1) between the ancient RNA-protein world and LUCA (the last universal common ancestor); and 2) between LUCA and LECA (the last eukaryotic common ancestor). On Earth, life divides into three domains: bacteria, archaea and eukaryotes. 7
Information flow within living organisms requires genetic material in DNA to be read and deciphered. Because, within the double helix, DNA bases project inward, DNA is more stable than single-stranded RNA, in which bases are more exposed. LUCA, therefore, developed a more stable DNA genome than was possible in the RNA-protein world. The DNA double helix, however, is difficult to unzip, so proteins must act on DNA to separate strands. The relative security of a DNA genome compared to a fragmented RNA genome comes with conditions.
LUCA (~3.5 to 3.8 billion years ago) was one of the first organisms with a streamlined DNA genome. The Earth is only ~4.5 billion years old, so LUCA is very ancient and the time for the RNA-protein world that preceded LUCA was short. To read the genetic information within a DNA genome requires a mobile protein factory called RNA polymerase. To initiate RNA synthesis on a DNA template, RNA polymerases found in the three domains of life require protein factors that help them to recognize and open a promoter DNA sequence. In bacteria, sigma factors help RNA polymerases bind promoter DNA.
The evolutionary relationship, among bacterial sigma factors and archaeal TFB-TBP has not clearly been known.
Burton and Burton hypothesize that both sigma factors and TFB arose from a primordial initiation factor at LUCA with 4-HTH motifs, and bacteria and archaea diverged in evolution about the time of LUCA largely because of this difference in genome reading styles. Archaeal TFB lost 2-HTH motifs and gained compensating functions including cooperation with TBP. Because bacteria and archaea interpret (read; "transcribe") their DNA genomes differently to synthesize RNA, these organisms became distinct.
Question: how and why should that distinction have evolved ? the differences of transcriptions are hudge, eukaryotic cells use over 100 protein sub units.
Evolution of promoter DNA sequences has long been a mystery, but bacterial promoters somewhat resemble archaeal and eukaryotic promoters.
Its still today a mystery, and to the well intended its obvious why. The resemblance of one with the other means nothing.
very early in evolution, bacteria and archaea came to read and interpret genomic information in DNA in fundamentally different ways, causing bacteria and archaea to diverge into distinct domains.
Question: how did they " learn " that feat ??
At LECA (~1.6-2.5 billion years ago) an archaea fused with multiple bacteria to form the first eukaryote.
Cool story. How do they possibly know ? Answer : they don't. There are hudge differences between the two :
Gene expression is linked to RNA transcription, which cannot happen without RNA polymerase. However, this is where the similarities between prokaryote and eukaryote expression end. 8
Transcription in Bacteria
In bacteria, all transcription is performed by a single type of RNA polymerase. This polymerase contains four catalytic subunits and a single regulatory subunit known as sigma (s). Interestingly, several distinct sigma factors have been identified, and each of these oversees transcription of a unique set of genes. Sigma factors are thus discriminatory, as each binds a distinct set of promoter sequences.
Transcription in Eukaryotes
Eukaryotic cells are more complex than bacteria in many ways, including in terms of transcription.
Specifically, in eukaryotes, transcription is achieved by three different types of RNA polymerase
These polymerases differ in the number and type of subunits they contain, as well as the class of RNAs they transcribe; that is, RNA pol I transcribes ribosomal RNAs (rRNAs), RNA pol II transcribes RNAs that will become messenger RNAs (mRNAs) and also small regulatory RNAs, and RNA pol III transcribes small RNAs such as transfer RNAs (tRNAs). Because RNA pol II transcribes protein-encoding genes, it has been of particular importance to scientists who study the regulation of eukaryotic gene expression, and its function is well understood. For example, researchers know that RNA pol II can bind to a DNA sequence within the promoter of many genes, known as the TATA box, to initiate transcription.
So there are remarkable differences and a dramatic increase of complexity of this machinery between prokaryotic and eukaryotic cells. Its hard to see how and why there could have been the evolution from one to the other 3 in eukaryotic cells.
Together with other common motifs (short recognition sequences in the DNA), these elements constitute the core promoter.
So there is not only a difference between the RNA polymerase machinery, but also in the promoters sections in DNA.
Prokaryotic vs. Eukaryotic Trancription
Differences:
Prokaryotes only contain three different promoter elements: -10, -35 promoters, and upstream elements. Eukaryotes contain many different promoter elements: TATA box, initiator elements, downstream core promoter element, CAAT box, and the GC box to name a few. Eukaryotes have three types of RNA polymerases, I, II, and III, and prokaryotes only have one type. Eukaryotes form and initiation complex with the various transcription factors that dissociate after initiation is completed. There is no such structure seen in prokaryotes. Another main difference between the two is that transcription and translation occurs simultaneously in prokaryotes and in eukaryotes the RNA is first transcribed in the nucleus and then translated in the cytoplasm. RNAs from eukaryotes undergo post-transcriptional modifications including: capping, polyadenylation, and splicing. These events do not occur in prokaryotes. mRNAs in prokaryotes tend to contain many different genes on a single mRNA meaning they are polycystronic. Eukaryotes contain mRNAs that are monocystronic. Termination in prokaryotes is done by either rho-dependent or rho-independent mechanisms. In eukaryotes transcription is terminated by two elements: a poly(A) signal and a downstream terminator sequence. 9
These are hudge differences. How could or should the transition from one kind of promoters and RNA polymerase machinery have occured to the other domain, far more complex ?
The RNA in a cell is made by DNA transcription, a process that has certain similarities to the process of DNA replication. The machines doing this job, RNA polymerase enzymes, are essential to life and are found in all organisms and many viruses.
By selecting which RNAs are made, RNA polymerase (RNAP) dictates how cells adapt to new environments, interact symbiotically or pathogenically with hosts, respond to stress and starvation, and multiply. RNAP accomplishes this task by deciding where and how often to start transcription, how to elongate RNAs, and where to stop. 6
Question: How did the cell " learn " how to decide where, and how often to start transcription ? Had this mechanism not have to be in place before the first cell was ready to replicate ?
Emergence of transcription initiation factors in the three domains of life.
The bacterial RNA polymerase (RNAP) strictly depends on σ-factors for transcription initiation, whereas archaeal and eukaryotic RNAPs require the initiation factors TATA box-binding protein (TBP) and transcription factor IIB (TFIIB). There are four potential scenarios for the evolution of these factors. As the first scenario requires the fewest independent evolutionary events, it is the simplest explanation for the occurrence of transcription initiation factors in all extant life. a | As there are no σ-factor homologues in archaea and eukaryotes, and no TBP and TFIIB homologues in bacteria, it is likely that the ancestral RNAP of the last universal common ancestor (LUCA) used neither σ-factors nor TBP or TFIIB factors, and that these factors evolved independently in the bacterial and archaeal–eukaryotic lineages, respectively, after their split. b | An alternative scenario is that the RNAP of the LUCA used both σ-factors and TBP and TFIIB factors in parallel, and then lost the relevant factors in each lineage. c | A third scenario is that the LUCA used σ-factors, and then the archaeal–eukaryotic lineage lost these factors are gained TBP and TFIIB factors. d | The final scenario is that the LUCA used TBP and TFIIB factors, and that these were then lost in the bacterial lineage and σ-factors were gained.10

There are four potential scenarios for the evolution of these transcription factors. Translated : they have no idea how the different factors could have evolved , and the mechanisms involved in a detailled manner. They just make things up. If someone expected to read the paper on how this large, complex transcription machine evolved, you would be disappointed. The authors merely assume it evolved, and attempt to place various scenarios into an evolutionary tree where they don’t fit very well. This is typical of evolutionary scientific literature that already “knows” evolution is a “fact” but has no way to back up the claim. Highly-complex, interdependent systems that involves a coded , specified, complex language, translation, regulation, feedback, quality control and highest precision. The paper above fails completely to address the origin of the most efficient information-processing system known on earth. And so do all mainstream scientific papers. Their frequent answer is : " We don't know yet ". As if one day they would know, and evolution would be confirmed...... Wait a minute. Really ??
Bacterial RNA polymerase is a huge factory with many moving parts. It is composed of a dozen different proteins. Together, they form a machine that surrounds DNA strands, unwinds them, and builds an RNA strand based on the information held inside the DNA. Once the enzyme gets started, RNA polymerase marches confidently along the DNA copying RNA strands thousands of nucleotides long. 1
Observe the nomenclature : huge factory, machine, marches. Question : how did the dozens of proteins arise ? Unless this protein machine was not fully formed, no mRNA could be formed, no transcription and subsequently, no replication would be possible, upon which mutations and natural selection could act. So the only alternatives left are chance, physical necessity, and design. Could it be that chance, or physical necessity are good explanations for the arise of the enormously complex RNA polymerase machine?
Accuracy
As you might expect, RNA polymerase needs to be accurate in its copying of genetic information. To improve its accuracy, it performs a simple proofreading stepas it builds an RNA strand.
Question: unless enough transcription accuracy is achieved, the whole process would not be accurate enough, and the whole process would be compromised. How could it have got this amazing feature upon natural ways ? Isnt that far better explained through a intentional designer, who created this essential mechanism ?
The active site is designed to be able to remove nucleotides as well as add them to the growing strand. The enzyme tends to hover around mismatched nucleotides longer than properly added ones, giving the enzyme time to remove them.
Question : how did the machine arise this amazing ability ?
This process is somewhat wasteful, since proper nucleotides are also occasionally removed, but this is a small price to pay for creating better RNA transcripts. Overall, RNA polymerase makes an error about once in 10,000 nucleotides added, or about once per RNA strand created.
Question: How did it get such a amazing accuracy ?
To transcribe a gene accurately, RNA polymerase must recognize where on the genome to start and where to finish.
Question: Had RNA polymerase not to be able to recognize where to start right from the beginning, otherwise it would not be functional ? How did it learn that feat ?
The initiation of transcription is an especially important step in gene expression because it is the main point at which the cell regulates which proteins are to be produced and at what rate.
Question : had the cell not to be able right from the start of its operation to regulate which proteins had to be produced, and and what rate ? How did it " learn " that feat ? Unless it was able to do so right from the beginning, would chaos not be the result ? Would the cell not cease to function properly ?
The bacterial RNA polymerase core enzyme is a multisubunit complex that sythesizes RNA using a DNA template as a guide.
Question : Unless it uses a DNA template as a guide, would the whole process also not be functional ? Had it not have to be able to use the DNA template as a guide right from the start ?
A detachable subunit called sigma (s) factor associates with the core enzyme and assists it in reading the signals in the DNA that tell it where to begin transcribing
Sigma factors:
http://en.wikipedia.org/wiki/Sigma_factor

These proteins belong to a family of polypeptides and they have very interesting features and play important role in specific gene expression.
They associate with RNAP subunits and they are also responsible for identifying the correct promoter and positioning the RNAP on DNA in proper context for initiating transcription. Otherwise the RNAP without sig cannot function as an enzyme; it is like a driver without a rudder/conductor.
Sigma (s) is a dissociable subunit of RNAP. When s is bound to core, the resulting complex, called holoenzyme, can bind with high affinity to promoter sites. 3)
Without the sigma factor, RNA polymerase would bind loosely and randomly to DNA, but would not be able to start transcription. Therefore no proteins would be made and the cell would die. 4)
Question: the sigma factor alone is LIFE ESSENTIAL. Had it not have to be there fully functional and programmed to do its job right when life started ?
Core RNA polymerase in bacteria is a complex composed of an
two alpha subunits
one beta, one beta’
one omega subunit,
together they form the core structure.
The α-amino-terminal domains (α-NTDs) serve as a scaffold for complex assembly;
the α-carboxy-terminal domains (α-CTDs) and ω-subunit play regulatory roles during initiation.
The β- and β′-subunits jointly form the active site and make all the contacts to the nucleic acids.


image ru
The process of transcription can be divided into three stages:
1) Initiation of a new RNA chain (parts 1-4 of the picture below).
2) Elongation of the chain (parts 5&6 of the picture below).
3) Termination of transcription and release of the brand new RNA molecule (parts 6&7 of the picture below).

upload pic
Together, s factor and core enzyme are known as the RNA polymerase holoenzyme; this complex adheres only weakly to bacterial DNA when the two collide, and a holoenzyme typically slides rapidly along the long DNA molecule until it dissociates again. However, when the polymerase holoenzyme slides into a region on the DNA double helix called a promoter, a special sequence of nucleotides indicating the starting point for RNA synthesis, the polymerase binds tightly to this DNA.
Question: Is it not essential, that this special sequence of nucleotides is there in the DNA code right from the beginning, otherwise the right mRNA cannot be synthesized ? How did these special sequences get there naturally ? How did the polymerase " learn " to bind tightly to this DNA ? Had it not have to be able to bind tightly right from the beginning ? Are these not functions, that are essential, and if they are not fully functional right from the beginning, they do not function at all ?
The polymerase holoenzyme, through its s factor, recognizes the promoter DNA sequence by making specific contacts with the portions of the bases that are exposed on the outside of the helix step 1 in the Figure above. After the RNA polymerase holoenzyme binds tightly to the promoter DNA in this way, it opens up the double helix to expose a short stretch of nucleotides on each strand (step 2 )
Question: how did the holoenzyme " learn " to do this precise step in a functional and correct way ? Try and fail attempts would not permit the process to go ahead, and the whole process would be compromised. Had the holoenzyme not have to be functional and fully formed right from the beginning ?
With the DNA unwound, one of the two exposed DNA strands acts as a template for complementary base-pairing with incoming ribonucleotides, two of which are joined together by the polymerase to begin an RNA chain (step 3 )

Questions:
Had the incoming and exit movement of the DNA strand not have to be pre programmed ?
Had the enter and exit channel of DNA not have to be precisely designed, and be there right from the beginning ? There could be no function of a incompletely designed enzyme.
Had the incoming and exit movement of the ribonucleotides and newly formed mRNA strand not have to be pre programmed ?
Had the enter and exit channel of ribonucleotides and the mRNA strand not have to be precisely designed, and be there right from the beginning ?
Had the ribonucleotides not have to be readily availabe for recruitment right from the beginning ? Had the entry channel and the exit channel of the ribonucleotides to enter and leave the factory and being moved and released to the place where the action goes on not to be designed?
After the first ten or so nucleotides of RNA have been synthesized (a relatively inefficient process during which polymerase synthesizes and discards short RNA oligomers), the core enzyme breaks its interactions with the promoter DNA, weakens its interactions with s factor, and begins to move down the DNA, synthesizing RNA (steps 4 and 5 )
Chain elongation continues (at a speed of approximately 50 nucleotides/sec for bacterial RNA polymerases) until the enzyme encounters a second signal in the DNA, the terminator , where the polymerase halts and releases both the newly made RNA chain and the DNA template (step 7 ) After the polymerase core enzyme has been released at a terminator, it reassociates with a free s factor to form a holoenzyme that can begin the process of transcription again.
Termination signals induce rapid and irreversible dissociation of the nascent transcript from RNA polymerase. Terminators at the end of genes prevent unintended transcription into the downstream genes, whereas terminators in the upstream regulatory leader regions adjust expression of the structural genes in response to metabolic and environmental signals. Premature termination within an operon leads to potentially deleterious defects in the expression of the downstream genes, but also provides an important surveillance mechanism. 5
Question : had the terminator not have to be there right from the beginning, otherwise the new mRNA strand would not have the right end ?
The processes of transcription initiation and termination involve a complicated series of structural transitions in protein, DNA, and RNA molecules. Unless the the whole operational sequence of the transcription cycle is not fully functional right from the start, transcription cannot occur. Take away just one of the steps, and the process ceases to function. Is it feasable and imaginable, that the RNA polymerase enzyme with its complex transcription process requiring many parts and subunits could be the product of random natural processes ? Transcription is a irreducible process. Furthermore, without replication , there is no evolution. In order for replication to occur, the DNA polymerase and RNA polymerase machinery had to be fully operational right since the start of life, so the machinery had to exist prior mutations and natural selection could occur.
Core promotors have to be encoded in DNA, otherwise the RNA polymerase enzyme does not know where to start and end transcription. Without it, no life would exist. It is impossible that it got there by trial and error. Unless it was at the right place right from the start, life could not happen. Feel free to bring chance into the game or good luck. You are guillable to the extreme, doing so. Either its fully functional, or it does not function at all. Is that not strong evidence for intelligent design ? If not, what explanation of origin fits better the observation described here ? [/justify]
http://reasonandscience.heavenforum.org/t2016-the-rna-polymerase-enzyme-and-its-function-evidence-of-design#3391
1) http://www.rcsb.org/pdb/101/motm.do?momID=40
2) http://www.proteopedia.org/wiki/index.php/2e2i
3) http://www.researchgate.net/profile/John_Helmann/publication/228031784_Sigma_Factors_in_Gene_Expression/links/0fcfd506ac99963842000000.pdf
4) http://helicase.pbworks.com/w/page/17605688/Matthew-Hunter
5) http://www.nature.com/nrmicro/journal/v9/n5/abs/nrmicro2560.html
6) http://landick.wisc.edu/images/geszvain_landick.pdf
7) https://bmb.natsci.msu.edu/about/directory/faculty/zachary-f-burton/genesis-of-life-on-earth/
8 )http://www.nature.com/scitable/topicpage/rna-transcription-by-rna-polymerase-prokaryotes-vs-961
9) http://www.chem.uwec.edu/webpapers2006/sites/demlba/folder/provseuk.html
10) http://www.nature.com/nrmicro/journal/v9/n2/fig_tab/nrmicro2507_F8.html
11) http://storage.cloversites.com/makinglifecountministriesinc/documents/Intelligent%20Design.pdf
Last edited by Admin on Sat Jul 11, 2020 3:25 am; edited 38 times in total