https://reasonandscience.catsboard.com/t1849-dna-replication-of-prokaryotes
The Argument of the Original Replicator
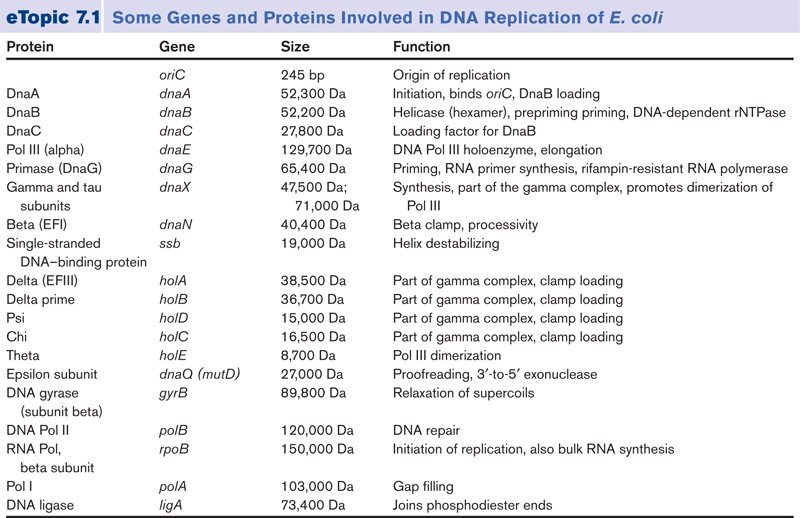
In prokaryotic cells, DNA replication involves more than thirty specialized proteins to perform tasks necessary for building and accurately copying the genetic molecule.
Each of these proteins is essential and required for the proper replicating process. Not a single one of these proteins can be missing, otherwise, the whole process breaks down, and is unable to perform its task correctly. DNA repair mechanisms must also be in place, fully functional, and working properly, otherwise, the mutation rate will be too high, and the cell dies. 18
The individual parts and proteins require by themselves complex assembly proteins to be built.
The individual parts, assembly proteins, and proteins individually would have no function on their own. They have only functions interconnected in the working whole.
The individual parts must be readily available on the construction site of the DNA replication complex, being correctly interlocked, interlinked, and have the right interface compatibility to be able to interact correctly together. All this requires information and meta-information ( information that directs the expression of the genomic information for construction of the individual proteins, and correct timing of expression, and as well the information of the correct assembly sequence. )
Evolution is not a capable driving force to make the DNA replicating complex, because evolution depends on cell replication through the very own mechanism we try to explain. It takes proteins to make DNA replication happen. But it takes the DNA replication process to make proteins. That’s a catch 22 situation.
DNA replication requires coded, complex, specified information and meta-information, and the DNA replication process is irreducibly complex.
Therefore, DNA replication is best explained through design.
Tan, Change; Stadler, Rob. The Stairway To Life:
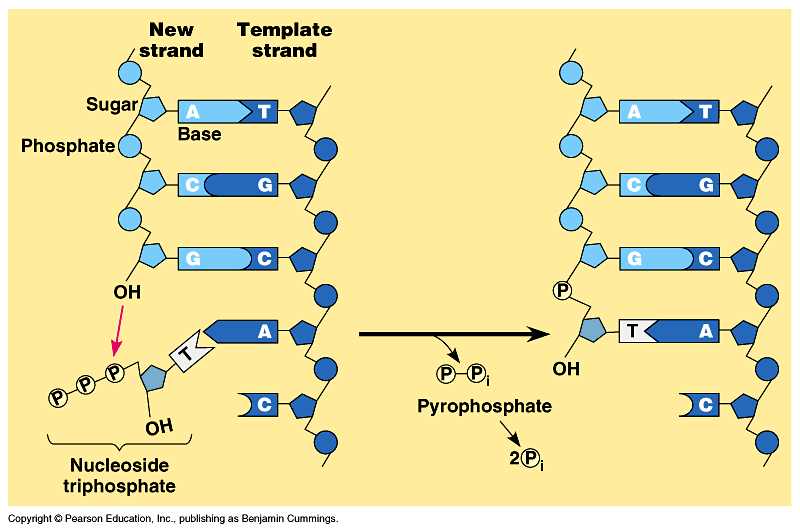
Combining the nucleotides
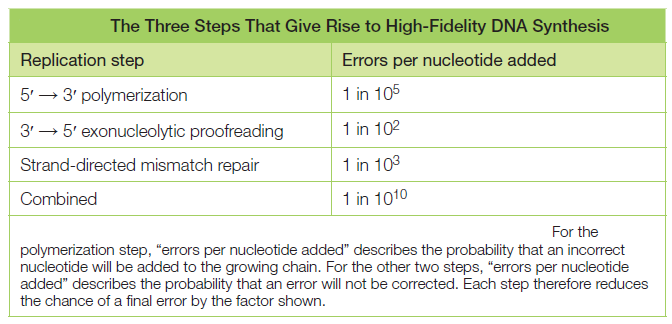
The natural replication process of DNA for E. coli has an error rate of approximately 1 in 1,000,000,000. It is not only incredibly accurate, but it is also incredibly fast (about one thousand nucleotides per second in E. coli). If DNA was scaled up to be one meter in diameter, the protein-based molecular machinery that replicates DNA “would move at approximately 600 km/hr (375 mph), and the replication machinery would be about the size of a FedEx delivery truck. Replicating the E. coli genome would be a 40 min, 400 km (250 mile) trip for two such machines, which would, on average make an error only once every 170 km (106 miles)”
Origin and Evolution of DNA and DNA Replication Machineries
The transition from the RNA to the DNA world was a major event in the history of life. The invention of DNA required the appearance of enzymatic activities for both synthesis of DNA precursors, retro-transcription of RNA templates and replication of single and double-stranded DNA molecules. Several of these enzymatic activities have been invented independently more than once, indicating that the transition from RNA to DNA genomes was more complex than previously thought. The distribution of the different protein families corresponding to these activities in the three domains of life (Archaea, Eukarya, and Bacteria) is puzzling. In many cases, Archaea and Eukarya contain the same version of these proteins, whereas Bacteria contain another version. However, in other cases, such as thymidylate synthases or type II DNA topoisomerases, the phylogenetic distributions of these proteins do not follow this simple pattern. Several hypotheses have been proposed to explain these observations, including independent invention of DNA and DNA replication proteins, ancient gene transfer and gene loss, and/or nonorthologous replacement.
https://www.ncbi.nlm.nih.gov/books/NBK6360/?fbclid=IwAR34sXRkv5xbOPBZEmSZhbB04SCYaY7cYsTOibuCSfH9aYC3NGohfWSEHCE
1. DNA Polymerase:
a. “DNA polymerases are spectacular molecular machines that can accurately copy genetic material with error rates on the order of 1 in 10^5 bases incorporated, not including the contributions of proofreading exonucleases.”
b. Part of the machine rotates 50° as the machine translocates along the DNA. These machines copy millions of base pairs of DNA every cell division so that each daughter cell gets an accurate copy.
c. “Although the polymerases are divided into several different families, they all share a common two metal-ion catalytic mechanism, and most of them are described as having fingers, palm, and thumb domains: the palm contains metal-binding catalytic residues, the thumb contacts DNA duplex, and the fingers form one side of the pocket surrounding the nascent base pair.” Three phases occur during each step along the DNA chain: the fingers open, the machine moves one base pair as it rotates, then the base in the “palm” is placed into the “pre-insertion site,” while another moving part prevents further movement till the operation is completed. Then the process repeats – millions of times per operation.
d. In no one of the articles describing DNA polymerase the word evolution was mentioned; no one can give this as an explanation.
The Argument of the Original Replicator
1. Evolution is the process by which an organism evolves from simpler ancestors.
2. Evolution by itself cannot explain how the original ancestor—the first living thing—came into existence (from 1).
3. The theory of natural selection can deal with this problem only by saying that the first living thing evolved out of non-living matter (from 2).
4. That original non-living matter (call it the Original Replicator) must be capable of:
(a) self-replication,
(b) generating a functioning mechanism out of surrounding matter to protect itself against falling apart, and
(c) surviving slight mutations to itself that will then result in slightly different replicators.
5. The Original Replicator is complex (from 4).
6. The Original Replicator is too complex to have arisen from purely physical processes (from 5 and The Classical Teleological Argument). For example, DNA, which currently carries the replicated design of organisms, cannot be the Original Replicator, because DNA molecules require a complex system of proteins to remain stable and to replicate, and could not have arisen from natural processes before complex life existed.
7. Natural selection cannot explain the origin and complexity of the Original Replicator (from 3 and 6).
8. The Original Replicator must have been created rather than have evolved (from 7 and The Classical Teleological Argument). Biologists and chemists have a theory with many ‘maybe’ which not everyone accepts as conclusive saying that it is theoretically possible for a simple physical system to make exact copies of itself from surrounding materials. Since then they have identified a number of naturally occurring molecules and crystals that can replicate in ways that could lead to natural selection (in particular, that allow random variations to be preserved in the copies). Once a molecule replicates, the process of natural selection can start creating, and the replicator can accumulate matter and become more complex, eventually leading to precursors of the replication system used by living organisms today.
9,"Unless the molecule can literally copy itself," Joyce and Orgel note, "that is, act simultaneously as both template and catalyst, it must encounter another copy of itself that it can use as a template." Copying any given RNA in its vicinity will lead to an error catastrophe, as the population of RNAs will decay into a collection of random sequences. But to find another copy of itself, the self-replicating RNA would need (Joyce and Orgel calculate) a library of RNA that "far exceeds the mass of the earth."In the face of these difficulties, they advise, one must reject the myth of a self-replicating RNA molecule that arose de novo from a soup of random polynucleotides. Not only is such a notion unrealistic in light of our current understanding of prebiotic chemistry, but it should strain the credulity of even an optimist's view of RNA's catalytic potential. If you doubt this, ask yourself whether you believe that a replicase ribozyme would arise in a solution containing nucleoside 5'-diphosphates and polynucleotide phosphorylase!
10. Where you get the idea that from matter life is possible. Evolutionist: "In the future we will do it." But in the original condition you show something. Just like formerly they were flying balloons. So because they were flying, they could say that "in the future we shall fly a big city– a Boeing 747." And in the history we can see that that is not impossible, because in the beginning condition or initiative condition we see that big things can be flown. But here and now you cannot even prepare an ant. You have not been able to prepare even a small ant, germ. Show me. So why do you say, "In future I shall do it"?
11. Anything that was created requires a Creator.
12. God exists.
For a nonliving system, questions about irreducible complexity are even more challenging for a totally natural non-design scenario, because natural selection — which is the main mechanism of Darwinian evolution — cannot exist until a system can reproduce. 17 For an origin of life we can think about the minimal complexity that would be required for reproduction and other basic life-functions. Most scientists think this would require hundreds of biomolecular parts, not just the five parts in a simple mousetrap or in my imaginary LMNOP system. And current science has no plausible theories to explain how the minimal complexity required for life (and the beginning of biological natural selection) could have been produced by natural process before the beginning of biological natural selection.
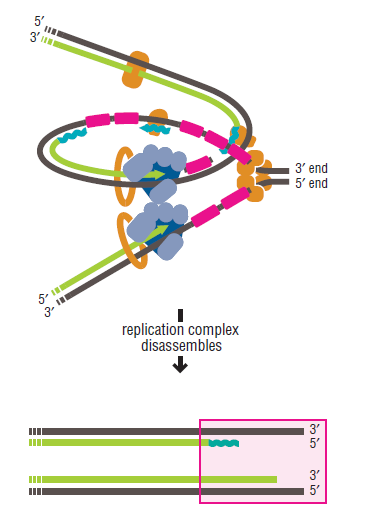
DNA replication is the most crucial step in cellular division, a process necessary for life, and errors can cause cancer and many other diseases. Genome duplication presents a formidable enzymatic challenge, requiring the high fidelity replication of millions of bases of DNA. It is a incredible system involving a city of proteins, enzymes, and other components that are breathtaking in their complexity and efficiency.
How do you get a living cell capable of self-reproduction from a “protein compound … ready to undergo still more complex changes”? Dawkins has to admit:
“Darwin, in his ‘warm little pond’ paragraph, speculated that the key event in the origin of life might have been the spontaneous arising of a protein, but this turns out to be less promising than most of Darwin’s ideas. … But there is something that proteins are outstandingly bad at, and this Darwin overlooked. They are completely hopeless at replication. They can’t make copies of themselves. This means that the key step in the origin of life cannot have been the spontaneous arising of a protein.” (pp. 419–20)
The process of DNA replication depends on many separate protein catalysts to unwind, stabilize, copy, edit, and rewind the original DNA message. In prokaryotic cells, DNA replication involves more than thirty specialized proteins to perform tasks necessary for building and accurately copying the genetic molecule. These specialized proteins include DNA polymerases, primases, helicases, topoisomerases, DNA-binding proteins, DNA ligases, and editing enzymes. DNA needs these proteins to copy the genetic information contained in DNA. But the proteins that copy the genetic information in DNA are themselves built from that information. This again poses what is, at the very least, a curiosity: the production of proteins requires DNA, but the production of DNA requires proteins.
Proponents of Darwinism are at a loss to tell us how this marvelous system began. Charles Darwin's main contribution, natural selection, does not apply until a system can reproduce all its parts. Getting a reproducible cell in a primordial soup is a giant leap, for which today's evolutionary biologists have no answer, no evidence, and no hope. It amounts to blind faith to believe that undirected, purposeless accidents somehow built the smallest, most complex, most efficient system known to man.
Several decades of experimental work have convinced us that DNA synthesis and replication actually require a plethora of proteins.
Replication of the genetic material is the single central property of living systems. Dawkins provocatively claimed that organisms are but vehicles for replicating and evolving genes, and I believe that this simple concept captures a key aspect of biological evolution. All phenotypic features of organisms—indeed, cells and organisms themselves as complex physical entities—emerge and evolve only inasmuch as they are conducive to genome replication. That is, they enhance the rate of this process, or, at least, do not impede it.
DNA replication is an enormously complex process with many different components that interact to ensure the faithful passing down of genetic components that interact to ensure the faithful passing down of genetic information to the next generation. A large number of parts have to work together to that end. In the absence of one or more of a number of the components, DNA replication is either halted completely or significantly compromised, and the cell either dies or becomes quite sick. Many of the components of the replication machinery form conceptually discrete sub-assemblies with conceptually discrete functions.
Wiki mentions that a key feature of the DNA replication mechanism is that it is designed to replicate relatively large genomes rapidly and with high fidelity. Part of the cellular machinery devoted to DNA replication and DNA-repair. The regulation of DNA replication is a vital cellular process. It is controlled by a series of mechanisms. One point of control is by modulating the accessibility of replication machinery components ( called the replisome ) to the single origin (oriC) region on the DNA. DNA replication should take place only when a cell is about to divide. If DNA replication occurs too frequently, too many copies of the bacterial chromosome will be found in each cell. Alternatively, if DNA replication does not occur frequently enough, a daughter cell will be left without a chromosome. Therefore, cell division in bacterial cells must be coordinated with DNA replication.
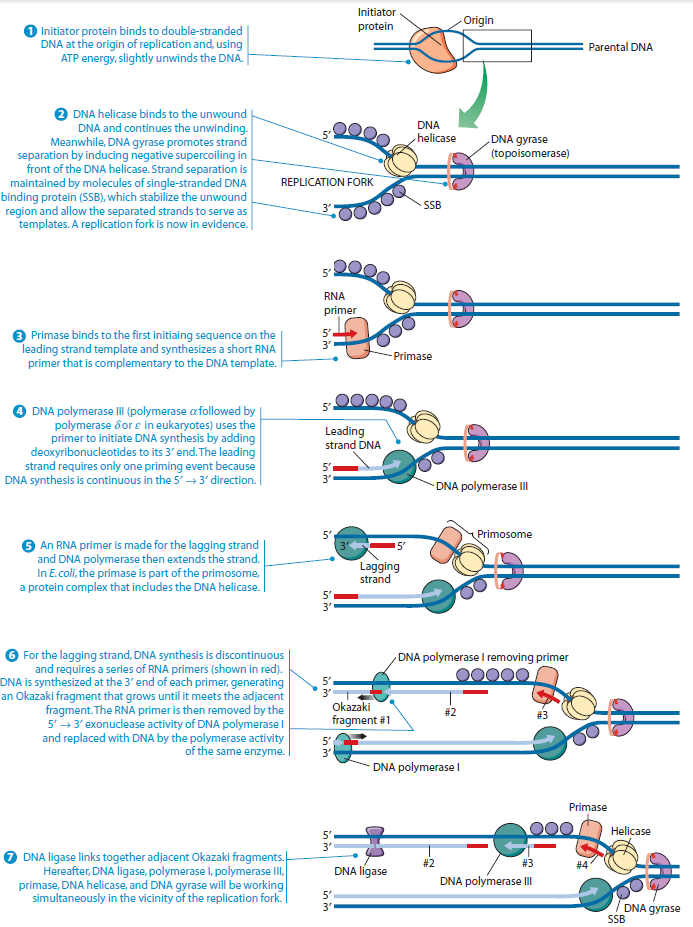
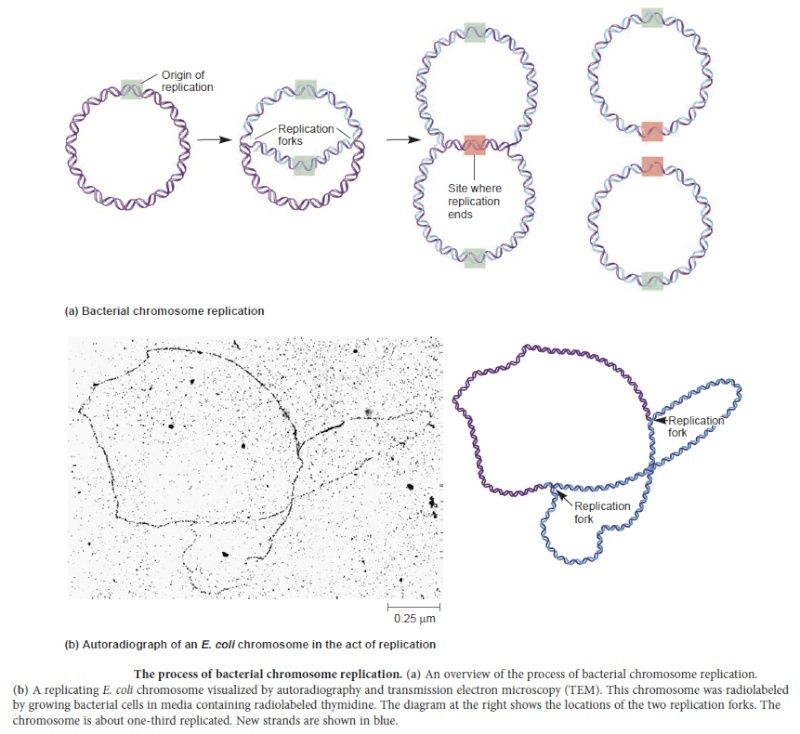
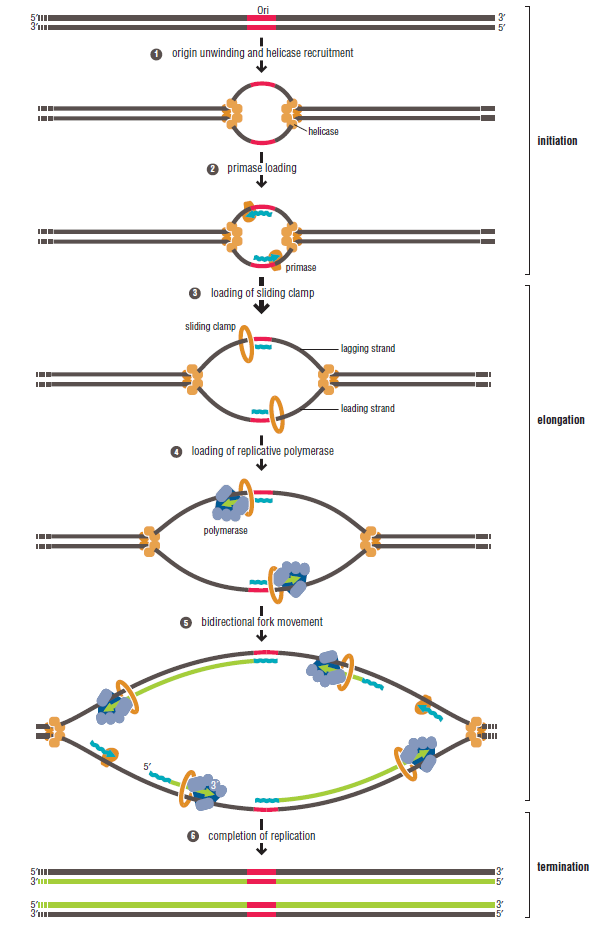
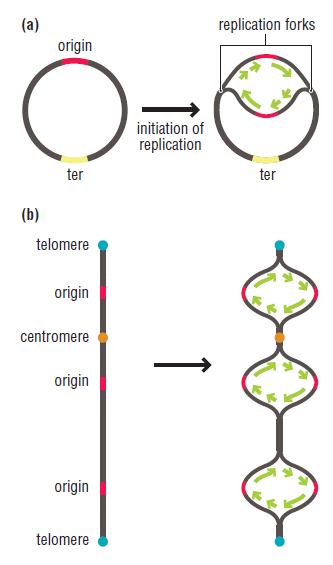
In prokaryotes, the DNA is circular. Replication starts at a single origin (ori C) and is bi-directional. The region of replicating DNA associated with the single origin is called a replication bubble and consists of two replication forks moving in opposite direction around the DNA circle. During DNA replication, the two parental strands separate and each acts as a template to direct the enzyme catalysed synthesis of a new complementary daughter strand following the normal base pairing rule. At least 10 different enzymes or proteins participate in the initiation phase of replication. Three basic steps involved in DNA replication are Initiation, elongation and termination, subdivided in eight discrete steps.
http://reasonandscience.heavenforum.org/t1849-dna-replication-of-prokaryotes#4365
Initiation phase:
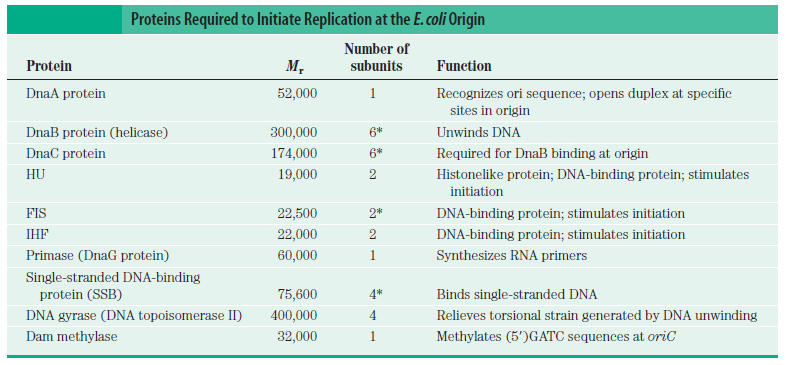
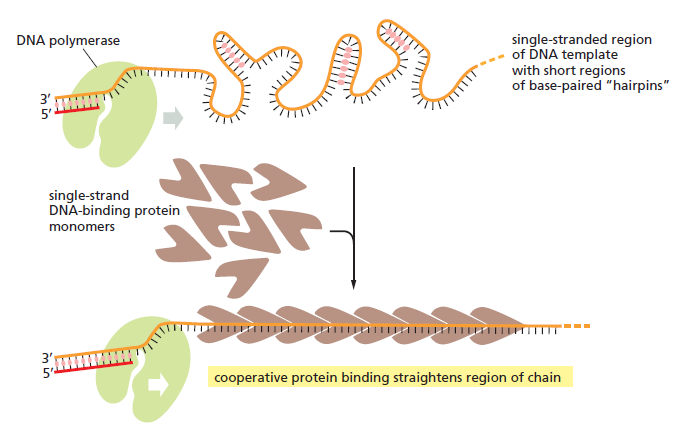
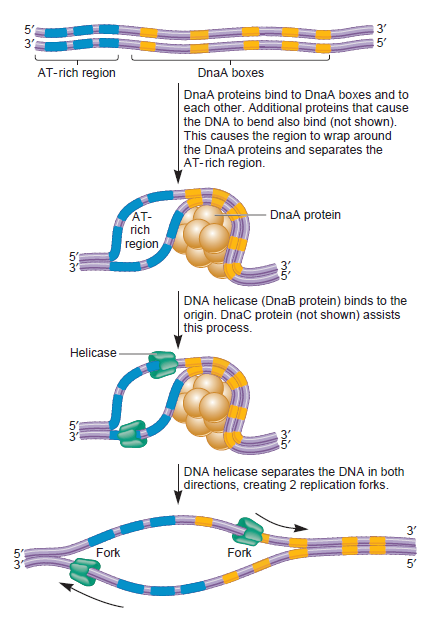
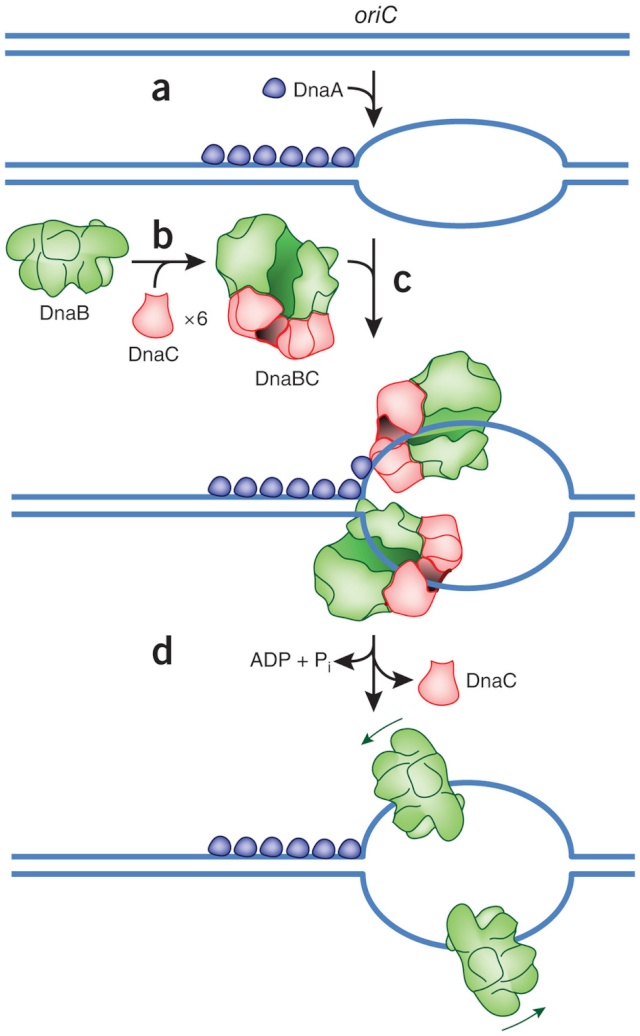
Step 1: Initiation begins, when DNA binds around an initiator protein complex DnaA with the goal to pull the two DNA strands apart. That creates a number of problems. First of all, the two strands like to be together - they stick to each other just as if they had tiny magnets up and down their length. In order to pull apart the DNA you have to put energy into the system. In modern cells, a protein called DnaA binds to a specific spot along the DNA, called single origin ( oriC ) and the protein proceeds to open up the double strand. The protein is a monomer, has motifs to bind to unique monomer sites, also they have motifs for protein-protein interaction, thus they can form clusters. They have hydrophobic regions for helical coiling and protein–protein interactions. Binding of the monomers to DnaA-A boxes, in ATP dependent manner (proteins have ATPase activity), leads to cooperative binding of more proteins. This clustering of proteins on DNA makes the DNA to wrap around the proteins, which induces torsional twist and it is this left handed twist that makes DNA to melt at 13-mer region and AT rich region; perhaps the negative super helical topology in this region may further facilitate the melting of the DNA. Opening or unwinding of dsDNA ( double strand DNA ) into single stranded region is an important event in initiation.
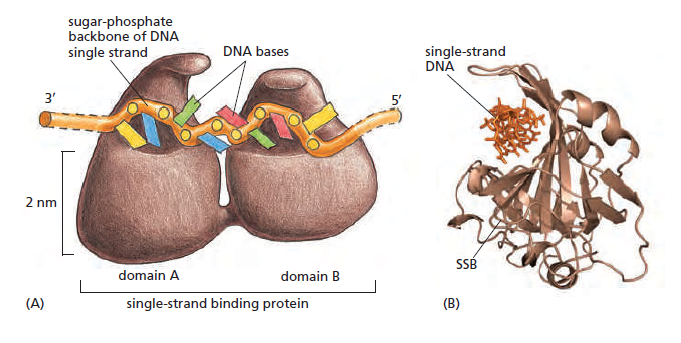
Single-strand binding protein (SSB)
https://reasonandscience.catsboard.com/t1849p15-dna-replication-of-prokaryotes#4377
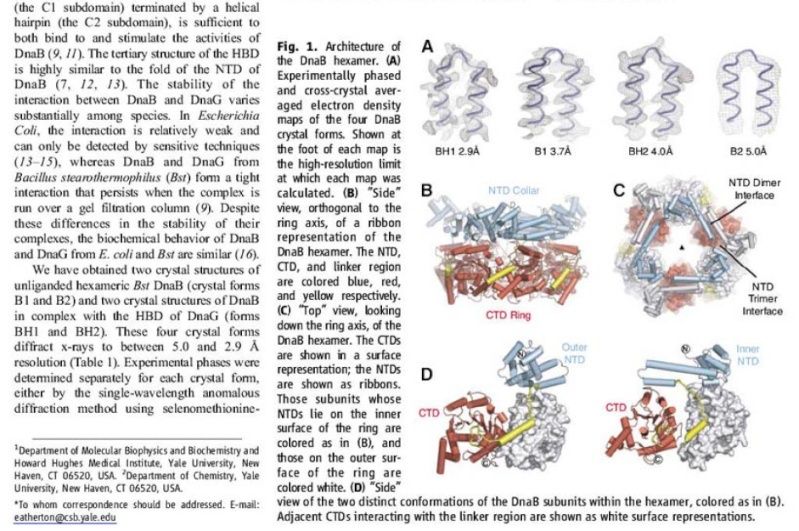
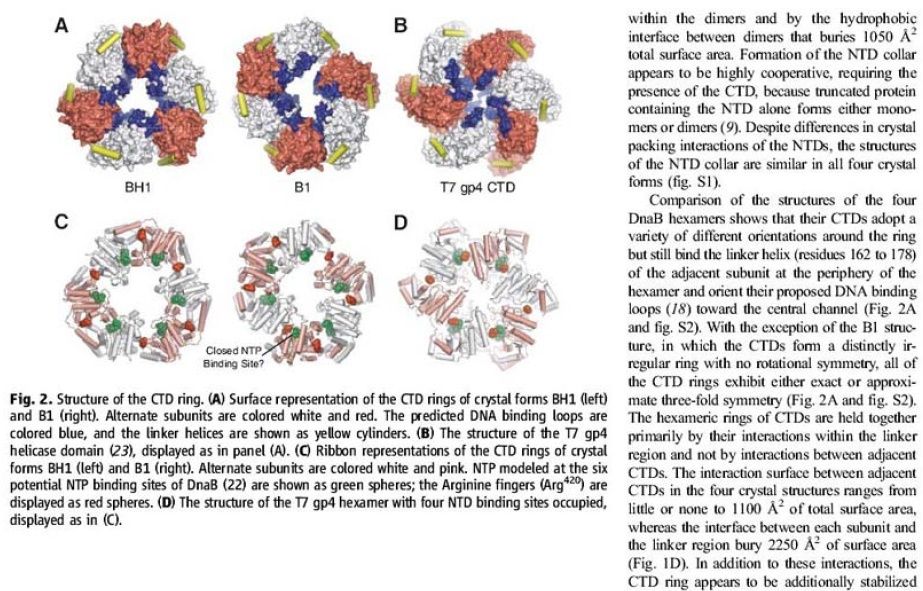
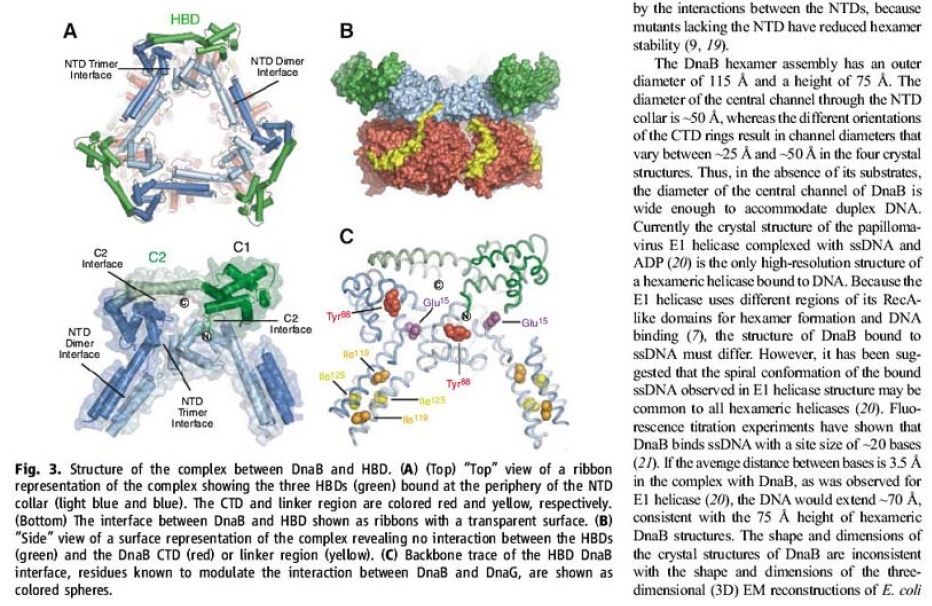
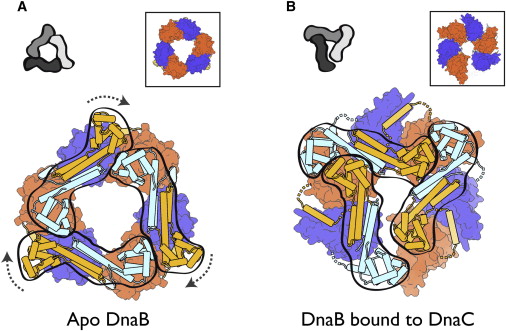
The Hexameric DnaB Helicase
https://reasonandscience.catsboard.com/t1849p15-dna-replication-of-prokaryotes#4367
DnaC, and strategies for helicase recruitment and loading in bacteria
https://reasonandscience.catsboard.com/t1849p15-dna-replication-of-prokaryotes#4371
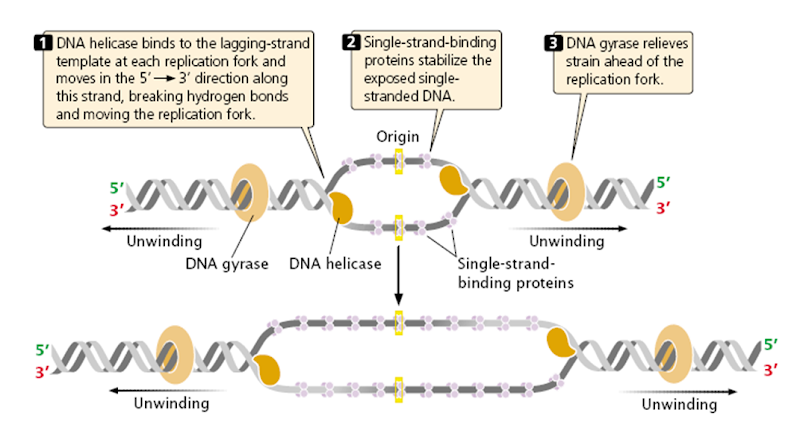
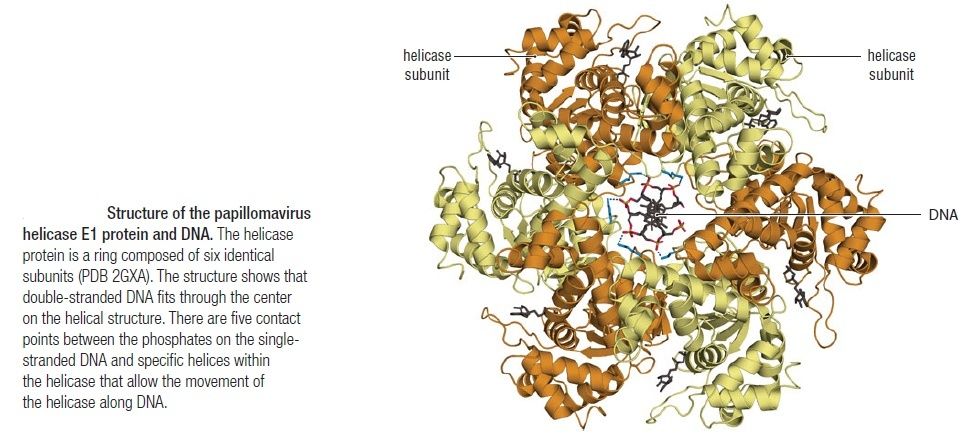
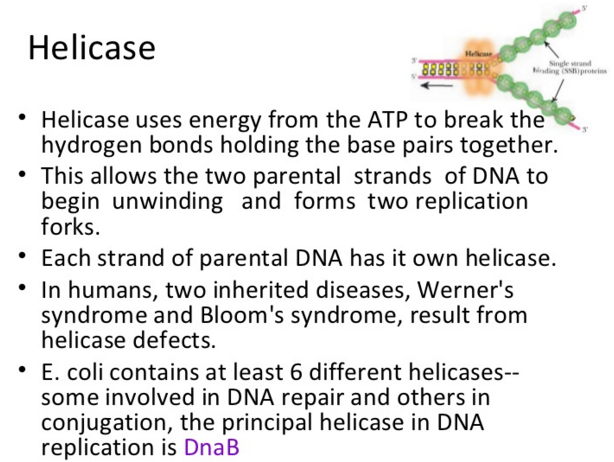
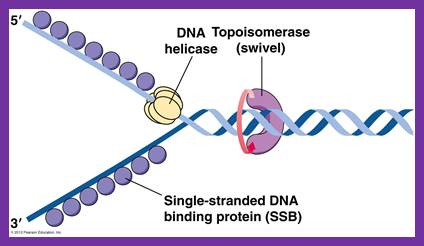
Unwinding the DNA Double Helix Requires DNA Helicases,Topoisomerases, and Single- Stranded DNA Binding Proteins
https://reasonandscience.catsboard.com/t1849p15-dna-replication-of-prokaryotes#4374
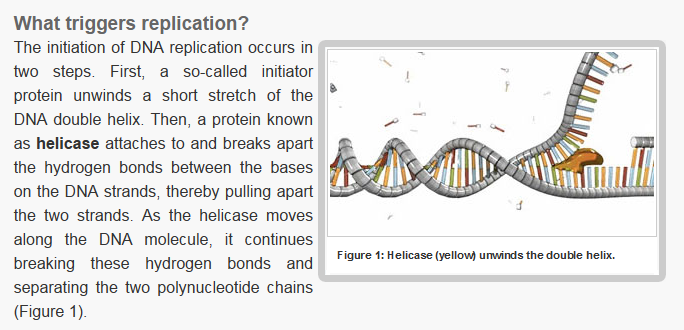
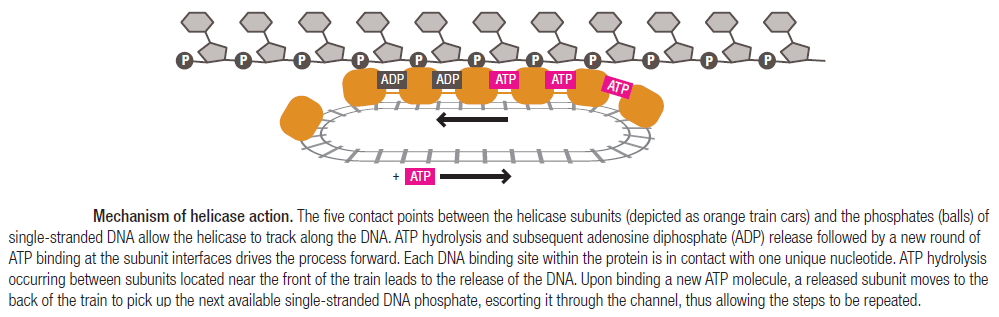
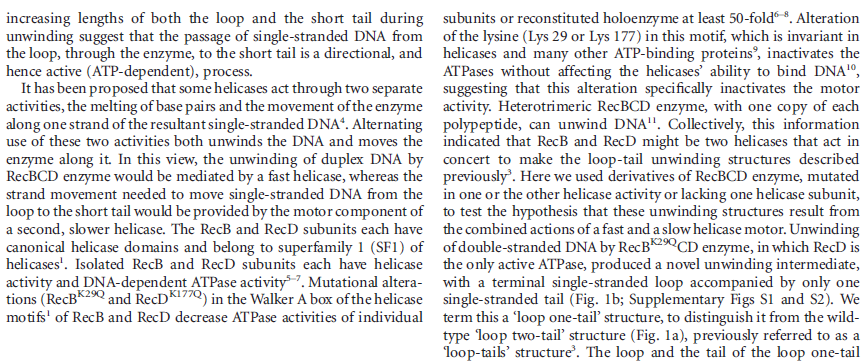
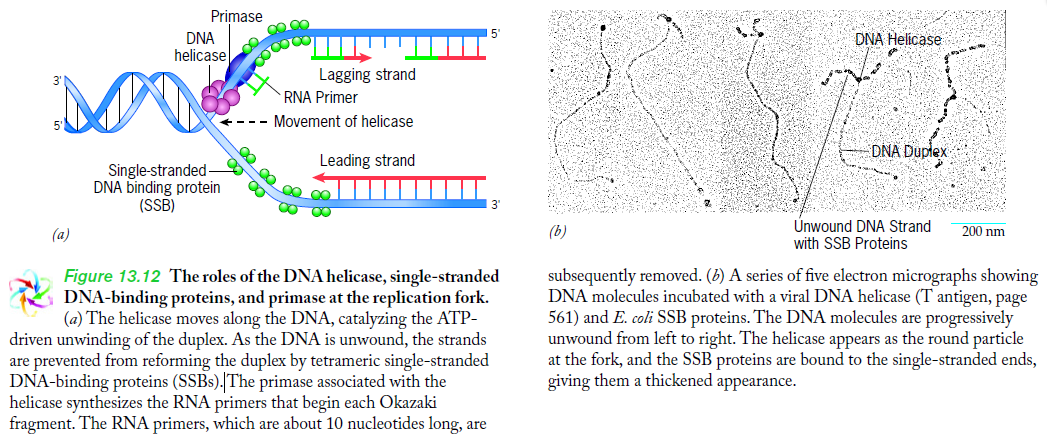
Step 2: During DNA replication, the two strands of the double helix must unwind at each replication fork to expose the single strands to the enzymes responsible for copying them. Three classes of proteins with distinct functions facilitate this unwinding process: DNA helicases, topoisomerases, and single-stranded DNA binding proteins ( SSB's). Helicase ( DnaB ) now comes along. The helicase exposes a region of single-stranded DNA that must be kept open for copying to proceed. Helicase is like a snowplow; it is a molecular machine that plows down the middle of the double helix, pushing apart the two strands. this allows the polymerase and associated proteins to travel along behind it in ease and comfort. DnaB helicase alone has no affinity for ssDNA ( single stranded DNA ) bound by SSB (single- stranded binding protein). Thus, entry of the DnaB helicase complex into the unwound oriC depends on DnaC, a additional protein factor. DnaC helps or facilitates the helicase to be loaded onto ssDNA at the replication fork in ATP dependent manner. The DnaB-DnaC complex forms a topologically open, three-tiered toroid. DnaC remodels DnaB to produce a cleft in the helicase ring suitable for DNA passage. DnaC’s fold is dispensable for DnaB loading and activation. DnaB possesses autoregulatory elements that control helicase loading and unwinding. Using energy derived from ATP hydrolysis, these proteins unwind the DNA double helix in advance of the replication fork, breaking the hydrogen bonds as they go. Helicase recruitment and loading in bacteria is a remarkable process. Following video shows how that works:
https://www.youtube.com/watch?v=YzNuLsqMqyE
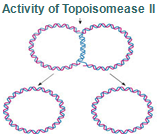
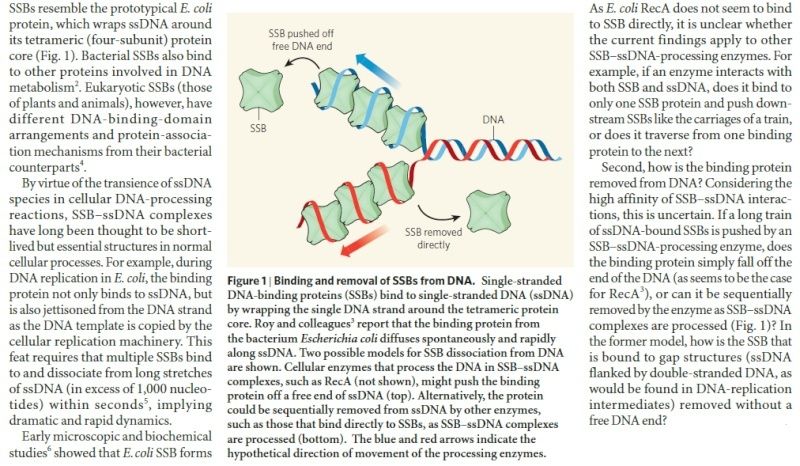
There is a problem, though, with this setup. If you push apart two DNA strands they generally do not float around separately. If they are close to one another they will rapidly snap back and form a double strand again almost as soon as the helicase passes. Even if the strands are not near each other, a single strand will usually fold up and form hydrogen bonds with itself - in other words, a tangled mess. So it is not enough to push apart the two strands of DNA; there must be a way to keep the strands apart once they have been separated. In modern cells this job is done by single-strand binding proteins, or SBB's. As the helicase separates the strands of DNA, SSB's bind to the single-stranded DNA and coats them. . SSB's prevent DNA from reannealing. SSB's associate to form tetramers around which the DNA is wrapped in a manner that significantly compacts the single-stranded DNA. There is another difficulty in being a double helix. The unwinding associated with DNA replication would create an intolerable amount of supercoiling and possibly tangling in the rest of the DNA. It can be illustrated with a simple example. Take two intertwined shoelaces and ask a friend to hold them together at each end. Now take a pencil, insert it between the strands near one end, and start pushing it down toward the other end. As you can see, shoestrings behind the pencil become melted, in the jargon of biochemistry. The shoestrings ahead of the pencil become more and more tangled. It becomes harder and harder to push the pencil forward. Helicase and polymerase encounter the same problem with DNA. It does not matter whether you are talking about interwined strings or interwined DNA strands. The problem of tangling is the result of the topological interconnectness of the two strands. If this problem persisted for very long in a cell, DNA replication would grind to a halt. However, the cell contains several enzymes, called topoisomerases, to take care of the difficulty. The way in which they do so can be illustrated with a enzyme called gyrase. Gyrase binds to DNA, pulls them apart and allows a separate portion of the DNA to pass through the cut. It then reseals the cut and lets go of the DNA. This action decreases the number of twists in DNA. The parental DNA is unwound by DNA helicases and SSB (travels in 5’-3’ direction), the resulting positive super-coiling (torsional stress) is relieved by topoisomerse I and II (DNA gyrase) by inducing transient single stranded breaks.Topoisomerases are amazing enzymes. In this topic, a video shows how they function :
Topoisomerase II enzymes, amazing evidence of design
https://reasonandscience.catsboard.com/t2111-topoisomerase-ii-enzymes-amazing-evidence-of-design?highlight=topoisomerase
In modern organisms, helicase, SSB, and gyrase all are required at the replication fork. Mutants in which any of them are missing are not viable - they die.
Question: Had not all three parts, the SSB binding proteins, the topoisomerase, and the helicase and the DnaC loading proteins not have to be there all at once, otherwise, nothing goes? They might exercise their function on their own, but then they would not replicate DNA or have function in a bigger picture. It's evident that they had to come together to provide a functional whole. What we see here are highly coordinated, goal-oriented tasks with specific movements designed to provide a specific outcome. Auto-regulation and control that seems required beside constant energy supply through ATP enhance the difficulty to make the whole mechanism work in the right manner. All this is awe-inspiring and evidences the wise guidance and intelligence required to make all this happening in the right way.
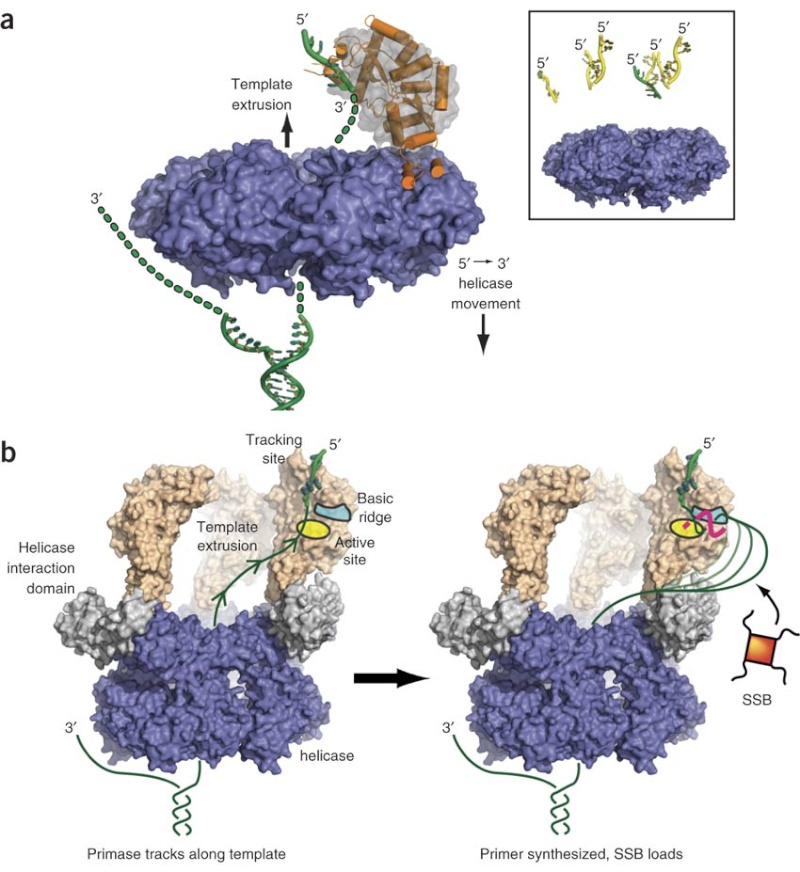
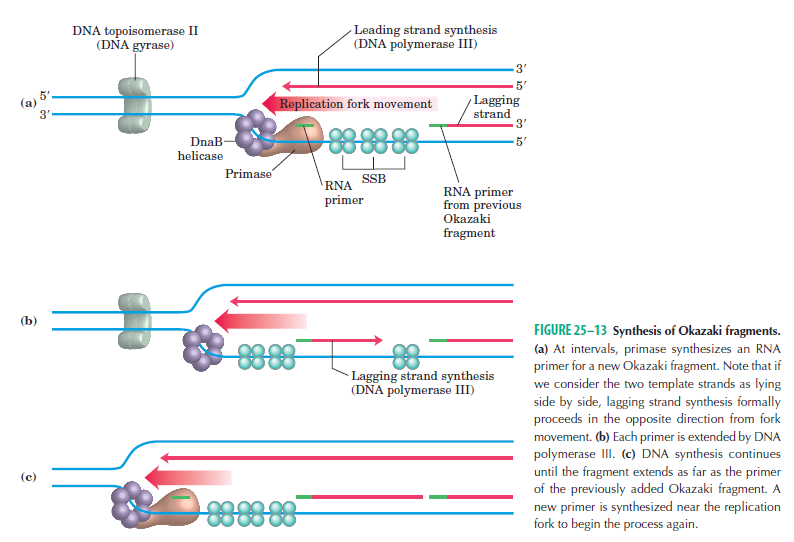
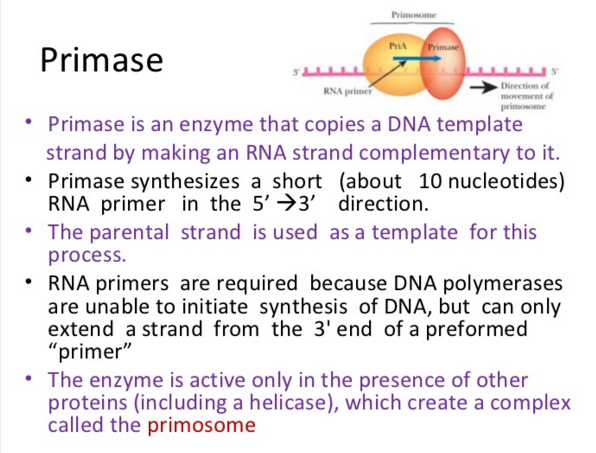
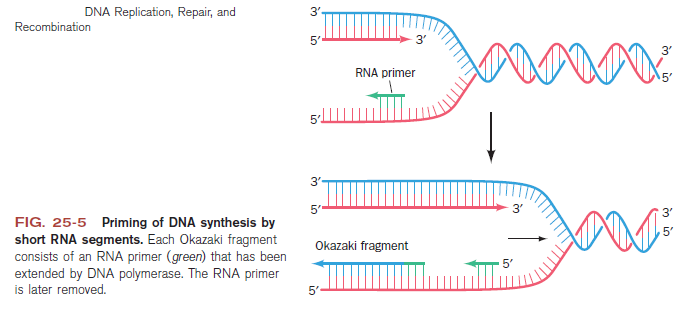
Step 3: The enzyme DNA primase (primase, an RNA polymerase) attaches to the DNA and synthesizes a short RNA primer to initiate synthesis of the leading strand of the first replication fork.
Elongation phase :
Step 4: In the elongation fase, DNA polymerase III extends the RNA primer made by primase.
DNA Polymerase
https://reasonandscience.catsboard.com/t1849p15-dna-replication-of-prokaryotes#4375
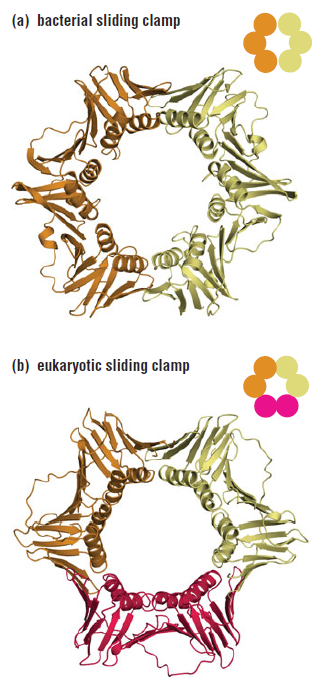
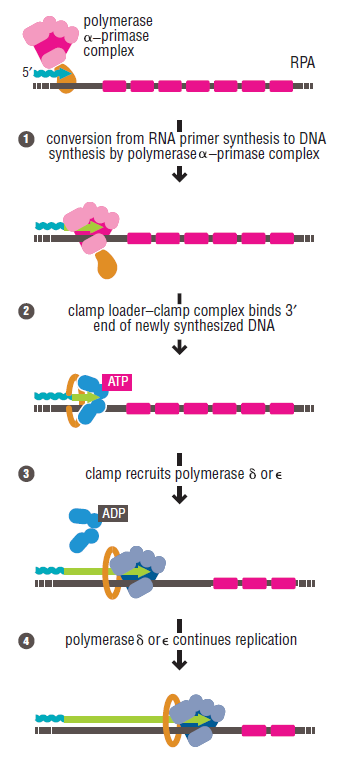
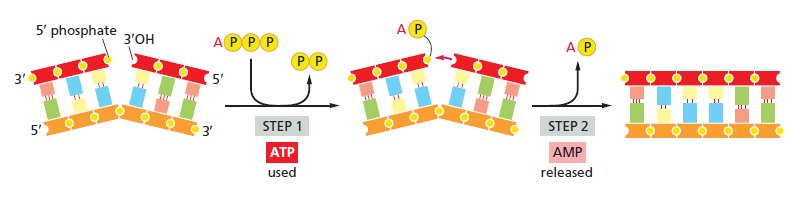
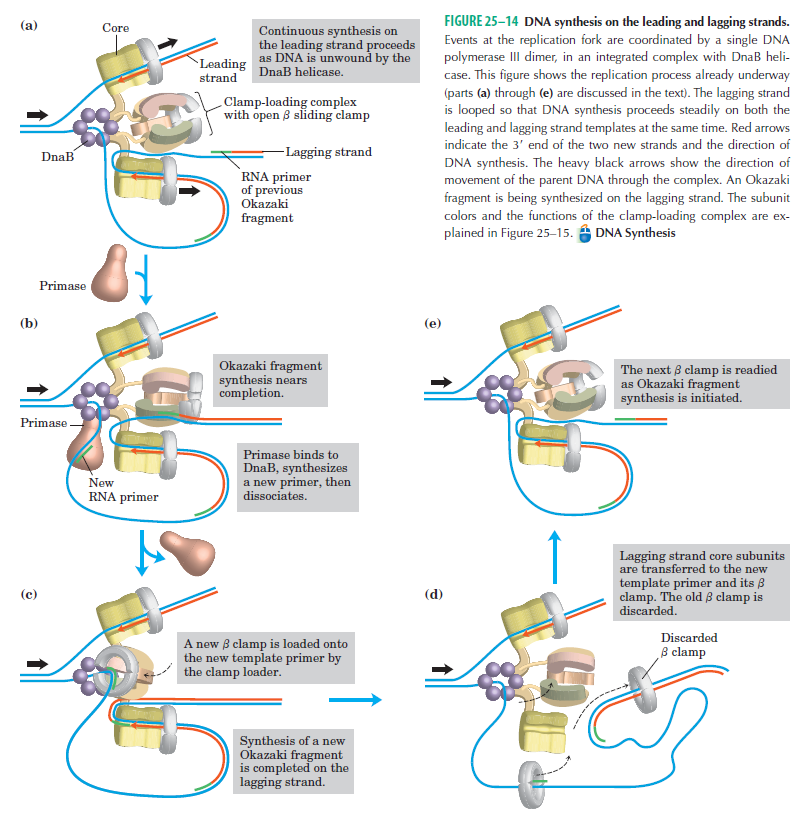
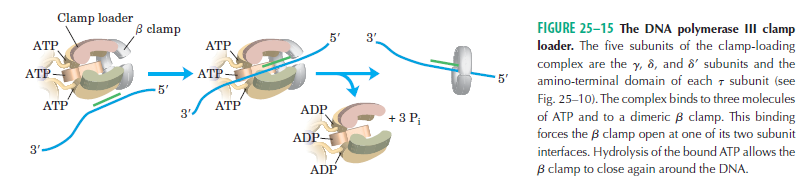
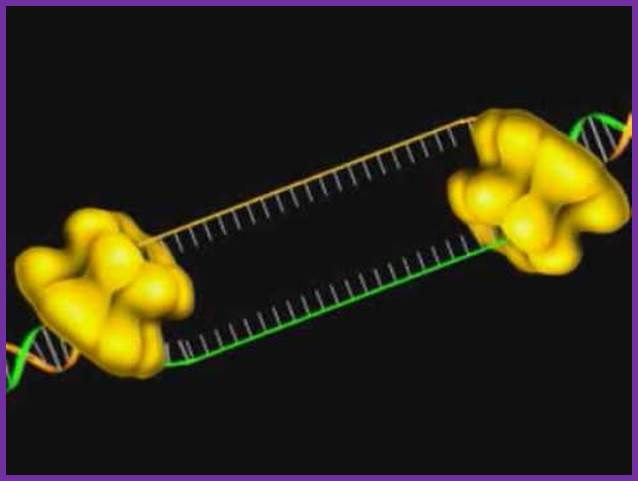
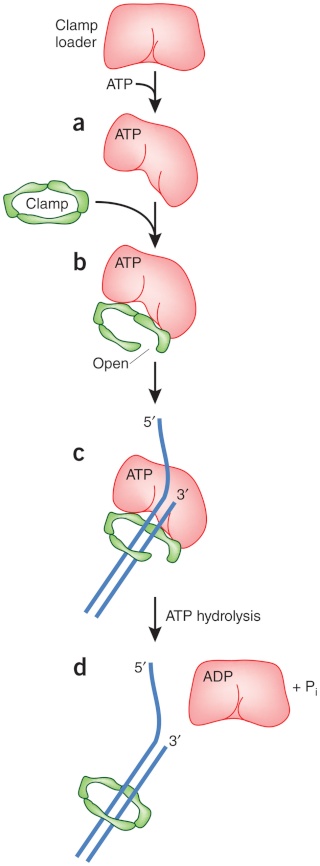
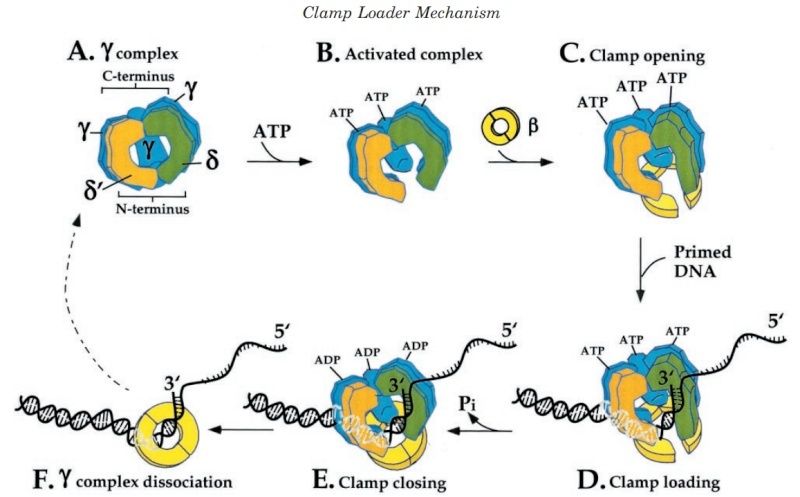
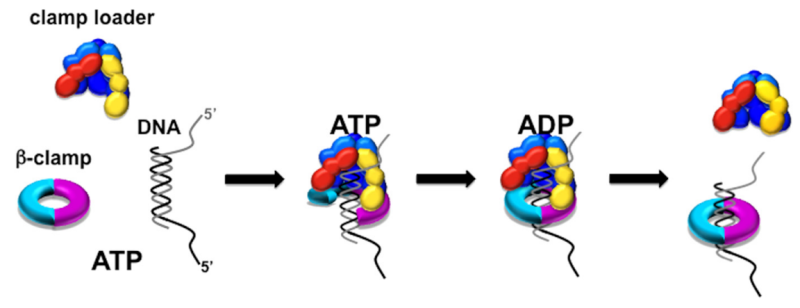
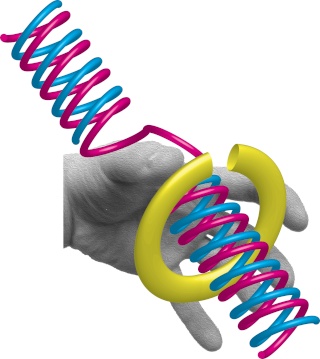
DNA polymerase possesses separate catalytic sites for polymerization and degradation of nucleic acid strands. All DNA polymerases make DNA in 5’-3’ direction . A ring-shaped sliding clamp protein encircles the DNA double helix and binds to DNA polymerase, thereby allowing the DNA polymerase to slide along the DNA while remaining firmly attached to it. Most enzymes work by colliding with their substrate, catalyzing a reaction and dissociating from the product. If that were the case with DNA polymerase, then it would bind to DNA, add a nucleotide to the new chain that was being made, and then fall off of the chain. Then ,put the next nucleotide onto the growing end, bind it and catalyze the addition. This same cycle would have to repeat itself a very large number of times to complete a new DNA chain. Polymerases however catalyze the addition of a nucleotide but do not fall off the DNA. Rather, they stay bound to it, until the next nucleotide comes in, and then they catalyze its addition to the chain. and they again stay bound. If it were not so, the replication process would be very slow. In the cell, polymerases stay on the DNA until their job is completed, which might be only after millions of nucleotides have been joined. This velocity is only possible because of clamp proteins. These have a ring shape. The ring can be opened up. These clamp proteins are joined to the DNA polymerase in a intricate way, through a clamp loader protein, which has a remarkable shape similar to a human hand. It takes the clamp, like a hand with five fingers would grab it, opens it up becoming like a doughnut shape,where the whole hole in the middle is big enough to accommodate the DNA, and then, when it is on the DNA, it positions it in a precise manner on the DNA polymerase, where it stays bound until it reaches the end of its polymerizing job. Through this ingenious process, the clamp stabilizes the DNA, making it possible to increase the speed of polymerization dramatically. They can be seen here:
The sliding clamp and clamp loader
https://reasonandscience.catsboard.com/t1849p15-dna-replication-of-prokaryotes#4376
Question: How would and could natural, unguided processes have figured out 1. the requirement of high-speed of polymerization? How could they have figured out the right configuration and process to do so? how could natural processes have emerged with the right proteins incrementally, with the hand-shaped clamp loader, and the precisely fitting clamp, enabling the fast process ?? Even the most intelligent scientists are still not able to imagine how this process is engineered? Furthermore, the process requires molecular energy in the form of ATP, and everything must fit together, and be functional. Without the clamp loader protein, the clamp could not be positioned to the polymerase enzyme, and processivity would not rise to the required speed. The whole process must also be regulated and controlled. How could that regulation have been programmed? Trial and error?
Several Proteins Are Required for DNA Replication at the Replication Fork
https://reasonandscience.catsboard.com/t1849p15-dna-replication-of-prokaryotes#4398
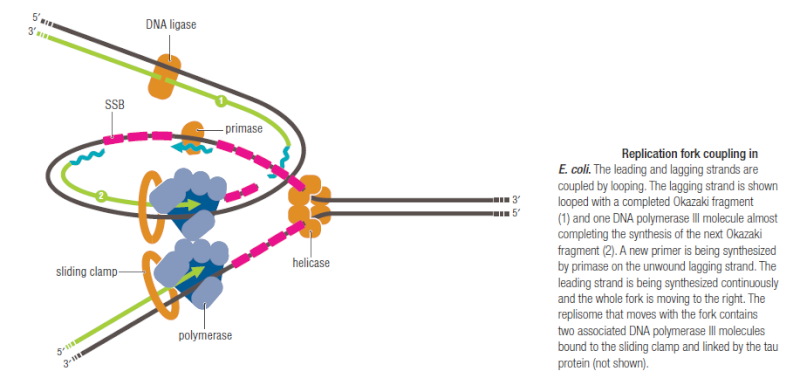
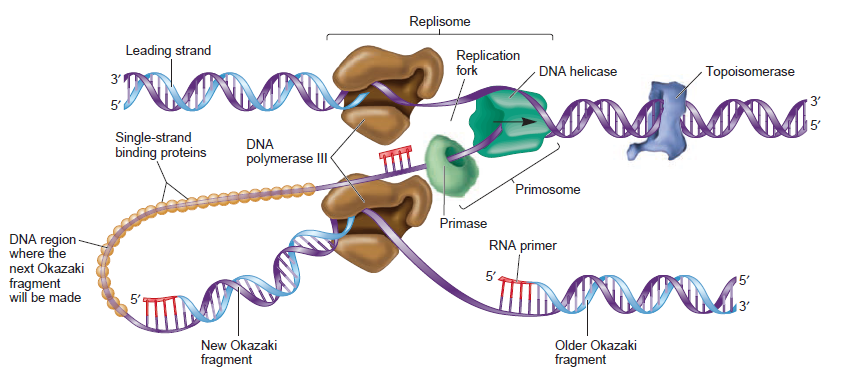
The various proteins involved in DNA replication are all closely associated in one large complex, called a replisome.
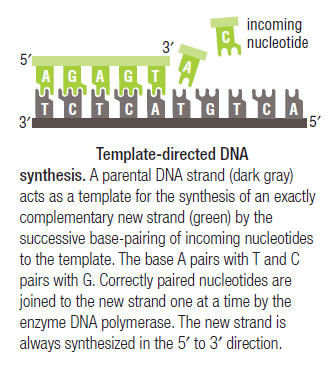
Leading strand synthesis: On the template strand with 3’-5’ orientation, new DNA is made continuously in 5’-3’ direction towards the replication fork. The new strand that is continuously synthesized in 5’-3’ direction is the leading strand.
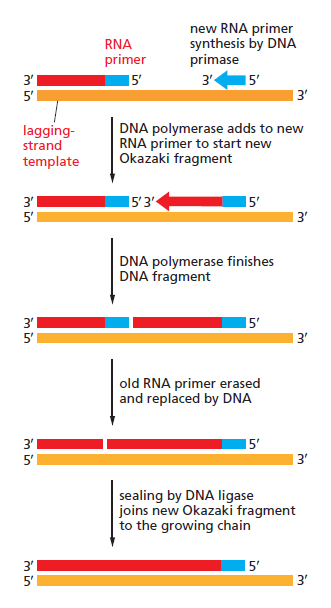
Lagging strand synthesis: In the lagging strand, the synthesis of DNA also elongates in a 5ʹ to 3ʹ manner, but it does so in the direction away from the replication fork. In the lagging strand, RNA primers must repeatedly initiate the synthesis of short segments of DNA; thus, the synthesis has to be discontinuous.
The Primase (DnaG) enzyme, and the primosome complex
https://reasonandscience.catsboard.com/t1849p15-dna-replication-of-prokaryotes#4379
The length of these fragments in bacteria is typically 1000 to 2000 nucleotides. In eukaryotes, the fragments are shorter—100 to 200 nucleotides. Each fragment contains a short RNA primer at the 5ʹ end, which is made by primase. The remainder of the fragment is a strand of DNA made by DNA polymerase III. The DNA fragments made in this manner are known as Okazaki fragments. To complete the synthesis of Okazaki fragments within the lagging strand, three additional events must occur: removal of the RNA primers, synthesis of DNA in the area where the primers have been removed, and the covalent attachment of adjacent fragments of DNA. In E. coli, the RNA primers are removed by the action of DNA polymerase I. This enzyme has a 5ʹ to 3ʹ exonuclease activity, which means that DNA polymerase I digests away the RNA primers in a 5ʹ to 3ʹ direction, leaving a vacant area. DNA polymerase I then synthesizes DNA to fill in this region. It uses the 3ʹ end of an adjacent Okazaki fragment as a primer. , DNA polymerase I would remove the RNA primer from the first Okazaki fragment and then synthesize DNA in the vacant region by attaching nucleotides to the 3ʹ end of the second Okazaki fragment. After the gap has been completely filled in, a covalent bond is still missing between the last nucleotide added by DNA polymerase I and the adjacent DNA strand that had been previously made by DNA polymerase III. To the left of the origin, the top strand is made continuously, whereas to the right of the origin it is made in Okazaki fragments. By comparison, the synthesis of the bottom strand is just the opposite. To the left of the origin it is made in Okazaki fragments and to the right of the origin the synthesis is continuous. Finally the two ends of the fragment have to be joined together; this is the job of an enzyme called DNA ligase. After the completion of one Okazaki fragment , the equipment has to be released, the clamp has to let go, and a new clamp has to be loaded at the beginning of the next fragment. Clearly the formation and control of the replication fork is an enormously complex process.
Step 5: After DNA synthesis by DNA pol III, DNA polymerase I uses its 5’-3’ exonuclease activity to remove the RNA primer and fills the gaps with new DNA. In the next step, finally DNA ligase joins the ends of the DNA fragments together. As the replisome moves along the DNA in the direction of the replication fork, it must accommodate the fact that DNA is being synthesized in opposite directions along the template on the two stands. Picture above provides a schematic model illustrating how this might be accomplished by folding the lagging strand template into a loop.Creating such a loop allows the DNA polymerase molecules on both the leading and lagging strands to move in the same physical direction, even though the two template strands are oriented with opposite polarity. The replisome faces special challenges as it makes new DNA at rates that can approach 1,000 nucleotides per second. Unlike the machines that make proteins and RNA, which work relatively sluggishly and in a linear fashion, the replisome must simultaneously copy two strands of DNA that are aligned in opposite directions (5ʹ to 3ʹ and 3ʹ to 5ʹ). Replisome chemistry obeys two rules.
Questions: How did they arise with that capability to " obey two rules " ? Suppose a primitive polymerase were duplicated and somehow started to replicate the second strand in the opposite direction while remaining attached to the first strand - how could that change have been directed, and why should that feat have happened randomly?

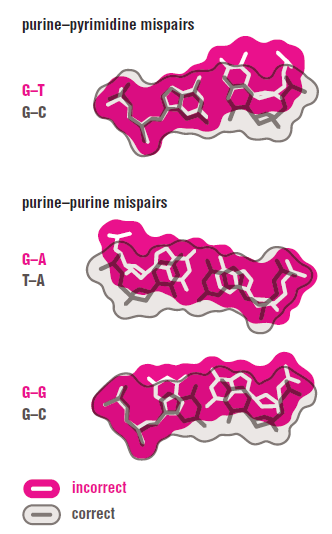
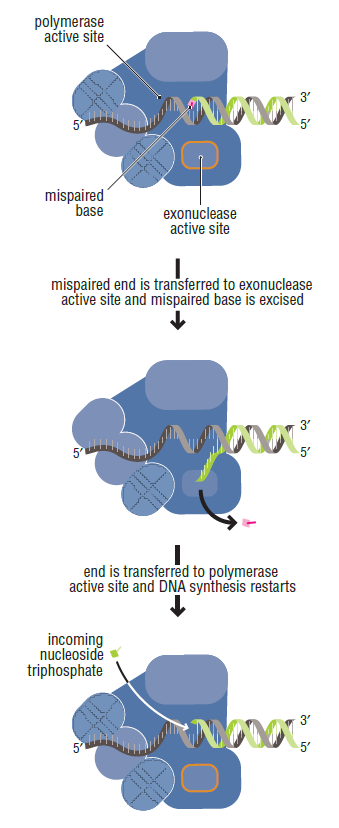
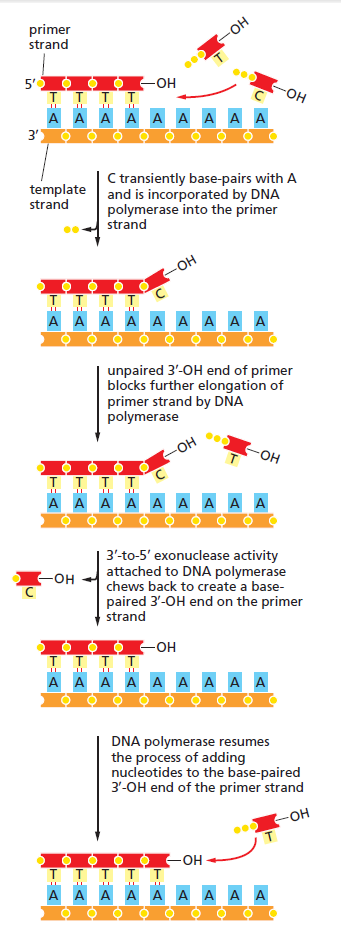
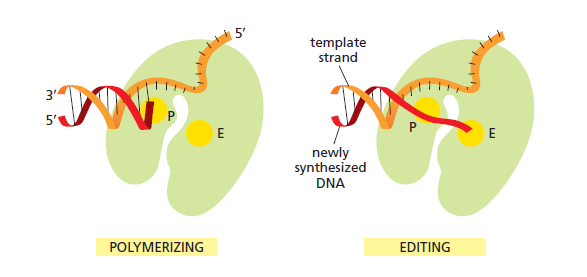
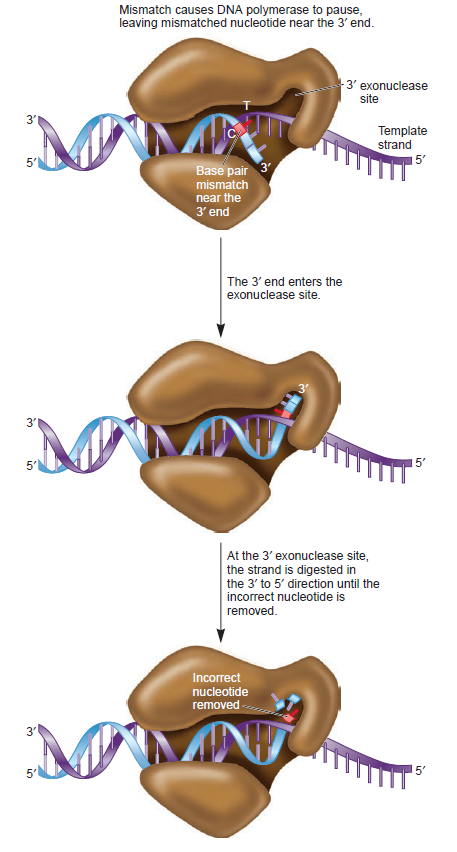
The DNA polymerase holoenzyme alone would not be able to duplicate the long DNA faithfully. Tests have shown that Polymerase III alone gets stuck. Furthermore, Polymerase III is not a simple enzyme. Its rather three enzymes in one. Beside replicating DNA, it can also degrade DNA in two different ways. It does so by three different, discrete regions of the molecule. The exonuclease activity plays a critical role in replication. It allows the enzyme to proofread the new DNA and cut out any mistakes it has made. Although the polymerase reads the sequence of the old DNA to produce a new DNA, it turns out that simple base bairing allows about one mistake per thousand base pairs copied. Proofreading reduces errors to about one mistake in a million base pairs. The question is if wheter a proofreading exonuclease and other DNA repair mechanisms had to be present in the very first cell.
Eigen’s theory revealed the existence of the fundamental limit on the fidelity of replication (the Eigen threshold): If the product of the error (mutation) rate and the information capacity (genome size) is below the Eigen threshold, there will be stable inheritance and hence evolution; however, if it is above the threshold, the mutational meltdown and extinction become inevitable (Eigen, 1971). The Eigen threshold lies somewhere between 1 and 10 mutations per round of replication (Tejero, et al., 2011) regardless of the exact value, staying above the threshold fidelity is required for sustainable replication and so is a prerequisite for the start of biological evolution. Indeed, the very origin of the first organisms presents at least an appearance of a paradox because a certain minimum level of complexity is required to make self-replication possible at all; high-fidelity replication requires additional functionalities that need even more information to be encoded (Penny, 2005). The crucial question in the study of the origin of life is how the Darwin-Eigen cycle started—how was the minimum complexity that is required to achieve the minimally acceptable replication fidelity attained? In even the simplest modern systems, such as RNA viruses with the replication fidelity of only about 10^3 and viroids that replicate with the lowest fidelity among the known replicons (about 10^2; Gago, et al., 2009), replication is catalyzed by complex protein polymerases. The replicase itself is produced by translation of the respective mRNA(s), which is mediated by the immensely complex ribosomal apparatus. Hence, the dramatic paradox of the origin of life is that, to attain the minimum complexity required for a biological system to start on the Darwin-Eigen spiral, a system of a far greater complexity appears to be required. How such a system could evolve is a puzzle that defeats conventional evolutionary thinking, all of which is about biological systems moving along the spiral; the solution is bound to be unusual.
DNA damage and repair
https://reasonandscience.catsboard.com/t2043-dna-repair?highlight=dna+repair
https://reasonandscience.catsboard.com/t1849p30-dna-replication-of-prokaryotes#4401
Replication forks may stall frequently and require some form of repair to allow completion of chromosomal duplication. Failure to solve these replicative problems comes at a high price, with the consequences being genome instability, cell death and, in higher organisms, cancer. Replication fork repair and hence reloading of DnaB may be needed away from oriC at any point within the chromosome and at any stage during chromosomal duplication. The potentially catastrophic effects of uncontrolled initiation of chromosomal duplication on genome stability suggests that replication restart must be regulated as tightly as DnaA-directed replication initiation at oriC. This implies reloading of DnaB must occur only on ssDNA at repaired forks or D-loops rather than onto other regions of ssDNA, such as those created by blocks to lagging strand synthesis.Thus an alternative replication initiator protein, PriA helicase, is utilized during replication restart to reload DnaB back onto the chromosome
Question: Could the first cell, with its required complement of genes coded for by DNA, have successfully reproduced for a significant number of generations without a proofreading function? A further question is how the function of synthesis of the lagging strand could have arisen, and the machinery to do so. That is, the Primosome, and the function of Polymerase I to remove the short peaces of RNA that the cell uses to prime replication, allowing the polymerase III function to fill the gap. These functions all require precise regulation, and coordinated functional machine-like steps. These are all complex, advanced functions and had to be present right from the beginning. How could this complex machinery have emerged in a gradual manner? the Primosome had to be fully functional, otherwise, polymerisation could not have started, since a prime sequence is required.
Step 6: Finally DNA ligase joins the ends of the DNA fragments together.
Termination phase:
Termination of DNA replication
https://reasonandscience.catsboard.com/t1849p15-dna-replication-of-prokaryotes#4399
Step 7: The two replication forks meet ~ 180 degree opposite to ori C, as DNA is circular in prokaryotes. Around this region there are several terminator sites which arrest the movement of forks by binding to the tus gene product, an inhibitor of helicase (Dna B).
Step 8: Once replication is complete, the two double stranded circular DNA molecules (daughter strands) remain interlinked. Topoisomerase II makes double stranded cuts to unlink these molecules.
The Astonishing Precision of DNA Replication
The astonishing accuracy and speed of DNA replication in organisms like E. coli underscore the remarkable efficiency of the molecular machinery involved in this essential biological process. Let's examine the implications of these statistics and the incredible capabilities of the replication machinery at a larger scale. With an error rate of approximately 1 in 1,000,000,000, DNA replication in E. coli achieves a level of fidelity that is unparalleled in human-made processes. This precision is a testament to the extremely accurate operating mechanisms and quality control systems in place during DNA synthesis. Such low error rates are crucial for maintaining the genetic integrity of an organism over countless generations. Moreover, the speed at which DNA replication occurs is equally remarkable. E. coli, a model organism for studying this process, can replicate at a rate of about one thousand nucleotides per second. When scaled up to human proportions, this would mean that the replication machinery, comprised of various enzymes and proteins, would be moving at an astounding speed of approximately 600 kilometers per hour (375 miles per hour). To put this into perspective, if we were to imagine DNA as a massive, one-meter-diameter circle, the replication machinery would be akin to a swift vehicle hurtling along at highway speeds. Visualizing this, it becomes clear that the replication machinery is not only swift but also highly efficient, as it manages to process and replicate DNA accurately at such high velocities. Now, consider the scenario where DNA is scaled up to such proportions that it is one meter in diameter. In this hypothetical scenario, the protein-based machinery responsible for DNA replication would be colossal, comparable in size to a FedEx delivery truck. This analogy underscores the complex and intricate nature of the molecular components involved in the replication process. Furthermore, let's contemplate the practical implications of this speed and accuracy. If we were to embark on a journey to replicate the entire E. coli genome, which consists of approximately 4.6 million base pairs, using this machinery, it would be a remarkably swift endeavor. The replication process would take a mere 40 minutes to complete a 400-kilometer (250-mile) journey. To put it in perspective, during this brief journey, these molecular machines, while moving at a breakneck pace, would only make an error in the genetic code once every 170 kilometers (106 miles). This astonishing level of precision allows organisms like E. coli to maintain their genetic information with incredible fidelity as they reproduce and pass their DNA on to future generations. The combination of extreme accuracy and rapidity in DNA replication in organisms like E. coli is a testament to the efficiency and sophistication of the molecular machinery involved. These attributes ensure the faithful transmission of genetic information, a fundamental requirement for the perpetuation of life on Earth.
According to mainstream scientific papers, the following twenty protein and protein complexes are essential for prokaryotic DNA replication. Each one mentioned below. They cannot be reduced. If one is missing, DNA replication cannot occur:
Pre-replication complex Formation of the pre-RC is required for DNA replication to occur
DnaA The crucial component in the initiation process is the DnaA protein
DiaA this novel protein plays an important role in regulating the initiation of chromosomal replication via direct interactions with the DnaA initiator.
DAM methylase It’s gene expression requires full methylation of GATC at its promoter region.
DnaB helicase Helicases are essential enzymes for DNA replication, a fundamental process in all living organisms.
DnaC Loading of the DnaB helicase is the key step in replication initiation. DnaC is essential for replication in vitro and in vivo.
HU-proteins HU protein is required for proper synchrony of replication initiation
SSB Single-stranded binding proteins Single-stranded DNA binding proteins are essential for the sequestration and processing of single-stranded DNA. 6
SSBs from the OB domain family play an essential role in the maintenance of genome stability, functioning in DNA replication, the repair of damaged DNA, the activation of cell cycle checkpoints, and in telomere maintenance. SSB proteins play an essential role in DNA metabolism by protecting single-stranded DNA and by mediating several important protein–protein interactions. 7
Hexameric DNA helicases DNA helicases are essential during DNA replication because they separate double-stranded DNA into single strands allowing each strand to be copied.
DNA polymerase I and III DNA polymerase 3 is essential for the replication of the leading and the lagging strands whereas DNA polymerase 1 is essential for removing of the RNA primers from the fragments and replacing it with the required nucleotides.
DnaG Primases They are essential for the initiation of such phenomena because DNA polymerases are incapable of de novo synthesis and can only elongate existing strands
Topoisomerases are essential in the separation of entangled daughter strands during replication. This function is believed to be performed by topoisomerase II in eukaryotes and by topoisomerase IV in prokaryotes. Failure to separate these strands leads to cell death.
Sliding clamp and clamp loader the clamp loader is a crucial aspect of the DNA replication machinery. Sliding clamps are DNA-tracking platforms that are essential for processive DNA replication in all living organisms
Primase (DnaG) Primases are essential RNA polymerases required for the initiation of DNA replication, lagging strand synthesis and replication restart. They are essential for the initiation of such phenomena because DNA polymerases are incapable of de novo synthesis and can only elongate existing strands.
RTP-Ter complex Ter sequences would not seem to be essential, but they may prevent overreplication by one fork in the event that the other is delayed or halted by an encounter with DNA damage or some other obstacle
Ribonuclease H RNase H1 plays essential roles in generating and clearing RNAs that act as primers of DNA replication.
Replication restart primosome Replication restart primosome is a complex dynamic system that is essential for bacterial survival.
DNA repair:
RecQ helicase In prokaryotes RecQ is necessary for plasmid recombination and DNA repair from UV-light, free radicals, and alkylating agents.
RecJ nuclease the repair machinery must be designed to act on a variety of heterogeneous DNA break sites.
Commentary: I am not aware of any scientific literature that provides a comprehensive, detailed explanation for the naturalistic emergence of DNA replication or any of its components de novo, without attributing a role to intelligent intervention. The intricate systems underpinning DNA replication surpass the capacity of unguided natural processes to explain them adequately. The high level of complexity and precision inherent in these mechanisms strongly suggests, in my perspective, the influence of an intelligent designer. The substantial coordination and specificity observed in these systems do not readily lend themselves to explanations rooted solely in chance or spontaneous generation, bolstering the argument for intelligent design as a credible and compelling explanation for the origin and mechanisms of DNA replication. Precisely because each of the detailed parts is essential, the entire system had to emerge simultaneously. Intelligence is known to have the capability to envision, design, and construct such motor-like systems based on extensive information. This knowledge further affirms that the emergence of such an intricate system through evolutionary processes is implausible. This is especially pertinent considering that the process of evolution itself inherently relies on the already established mechanism of DNA replication. Given these circumstances, it's rational to posit design as the most compelling explanation. The prospect of such a complex, interdependent system originating by chance is an untenable proposition. The individual components of the DNA replication system lack utility in isolation, and there's no plausible explanation for the spontaneous, random assembly of such an ordered, purposeful structure from disorder. It defies reason to assume that matter would arbitrarily organize itself into a highly specialized, complex, machine-like system, further reinforcing the argument for intelligent design as the most persuasive explanation for the origin of DNA replication.
Claim: DNA replication is due to millions of years of evolution, no god required!
Reply: DNA replication is a fundamentally complex process, essential for the propagation of life as it ensures the transmission of genetic information from one generation to the next. DNA replication had to be fully established at the onset of life and is grounded in the understanding that it is indispensable for cellular self-replication and subsequent evolutionary processes. Without a reliable mechanism to ensure accurate replication of genetic information, the continuity and diversification of life would be unattainable. The process of DNA replication in itself is a marvel of molecular coordination and precision. It involves a host of specialized proteins and enzymes that work in synchrony to unwind the DNA double helix, synthesize complementary strands, and correct errors to ensure the fidelity of the replicated DNA. For instance, the enzyme DNA polymerase plays a crucial role in synthesizing the new DNA strands, while helicase is responsible for unwinding the double helix, and ligase seals gaps to produce a continuous DNA strand.
The origin of this intricate system poses a substantial puzzle. How could such a complex, highly coordinated process spontaneously emerge in the primordial environment? The system's intricacy and the interdependence of its components render it a challenging scenario for gradual evolutionary assembly. Each component within the DNA replication machinery is integral, and the absence or malfunction of even a single element could lead to the collapse of the entire process. Moreover, DNA carries within its structure the coded instructions necessary for building and operating the organism. This information is fundamental for the synthesis of proteins, the workhorses of the cell responsible for virtually every task necessary for the cell’s structure, function, and regulation. The paradox here lies in the fact that the machinery required for reading and interpreting this genetic information (such as ribosomes and various RNA molecules) is itself encoded within the DNA. This creates a scenario akin to a “chicken and egg” problem: the information in DNA is necessary to produce the machinery that reads the information, yet this machinery is required to read and implement the information contained within the DNA.
The multifaceted and complex nature of DNA replication serves as a testimony to a purposeful creation by an intelligent source. The intricacies of the DNA replication process defy the expectations of a random emergence and autonomous assembly of life's fundamental machinery. The coordinated action of various proteins and enzymes such as DNA polymerase, helicase, and ligase, each performing a unique and indispensable function, reflects a system that is designed with precision and foresight. The level of coordination and complexity involved in DNA replication cannot be understated. The seamless interplay among various components to ensure the accurate duplication of genetic information exemplifies an orchestrated and purposeful operation, aligning more with deliberate design than with accidental emergence. The presence of multiple, interdependent components working concurrently to achieve a specific outcome – the accurate replication of DNA – resonates with the characteristics of designed systems observed in human-engineered technologies. Additionally, the information coding within the DNA molecule, essential for the construction and operation of living organisms, echoes the principles of intelligent communication systems. The precise, coded information embedded within the DNA sequence, and its role in dictating the synthesis of proteins, underscores the presence of a sophisticated information storage and retrieval system akin to advanced computational algorithms. This analogy bolsters the argument for intelligent design as the source of such a high-level information management system. The DNA replication system embodies irreducible complexity as it is composed of multiple components, each essential for the system's function. The absence of any single element would render the system nonfunctional, suggesting that the system must have been fully formed from the onset, contrary to the gradual assembly postulated by evolutionary mechanisms. The order and precision inherent in the DNA replication process further reinforce the argument for intelligent design. The specificity in the function of each component, the timing of each step, and the error-checking and correction mechanisms underscore a level of order and precision consistent with purposeful design rather than random occurrence. In view of these facts, the DNA replication process, with its complexity, coordination, information coding, irreducible complexity, and order and precision, stands as a compelling illustration of purposeful and intelligent design in the biological realm. They argue that such a sophisticated and intricate system, essential for life, more plausibly originates from an intelligent source than from unguided naturalistic processes.
https://www.youtube.com/watch?v=0Ha9nppnwOc
more: http://www.powershow.com/view1/214239-ZDc1Z/Biology_is_based_on_the_most_complex_chemical_systems_known_to_man_powerpoint_ppt_presentation
1) http://www.weizmann.ac.il/plants/Milo/images/MolMachinesSize120116Clean.pdf
2) http://cshperspectives.cshlp.org/content/5/10/a010116.full
3) https://en.wikipedia.org/wiki/Control_of_chromosome_duplication
4) http://www.biochem.umd.edu/biochem/kahn/molmachines/replication/Dna%20C%20protein.htm
5) http://www.nature.com/nsmb/journal/v15/n1/full/nsmb1356.html
6) http://www.biomedcentral.com/1471-2199/14/9
7) http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3632105/
8 ) Meyer, signature of the cell, page 111
9 ) http://journal.frontiersin.org/article/10.3389/fmicb.2014.00735/full#F1
10 ) http://creationsafaris.com/epoi_c08.htm
11) http://creationsafaris.com/ar_srds.htm
12) http://www.ncbi.nlm.nih.gov/books/NBK6360/
13) from the book: The Logic of Chance: The Nature and Origin of Biological Evolution By Eugene V. Koonin, page 266
14) https://www-als.lbl.gov/index.php/science-highlights/science-highlights/669-structures-of-clamp-loader-complexes-are-key-to-dna-replication.html
15) http://www.nature.com/nature/journal/v429/n6993/full/nature02585.html
16) http://ethos.bl.uk/OrderDetails.do?uin=uk.bl.ethos.604184
17) http://www.asa3.org/ASA/education/origins/ic-cr.htm
18) Inaccurate replication would likely have limited the size of the progenote genome due to the risk of “error catastrophe,” the accumulation of so many genetic mistakes that the organism is no longer viable. To illustrate this point, consider the problem of replicating a genome of one million bases, which is sufficient to encode a few hundred RNAs and proteins. (The smallest known genome for an extant free-living bacterium is that of Pelagibacter ubique, which consists of 1.3 million bases.) If replication were even modestly faithful, with an error frequency of 0.1%, every replication of a genome consisting of 1 million bases would result in 1000 errors, approximately one or two in every gene. Some of those errors would have been harmless, and a few might have been beneficial, but many would have been detrimental, leading to macromolecules with impaired functions.
further readings:
Resources for CHAPTER 3: REPLICATION : http://users.path.ox.ac.uk/~pcook/w1/3Replication.html#The_mechanics_of_synthesis_at_the_fork
Last edited by Otangelo on Fri Sep 29, 2023 6:26 am; edited 125 times in total
















































































 jstat
jstat