10.9. Key Cofactors in C1 Metabolism of Chemolithoautotrophs
Chemolithoautotrophs, which obtain energy from inorganic substances and carbon from CO2, are studied separately as potential models for early life forms due to their unique metabolic pathways and ability to thrive in extreme conditions. Their specialized enzymes, such as carbon monoxide dehydrogenase/acetyl-CoA synthase and hydrogenases, offer insights into primitive biochemical processes that could have existed in early Earth environments like hydrothermal vents. The study of one-carbon metabolism in both widespread and chemolithoautotrophic pathways provides a comprehensive approach to understanding the possible origins of life. In chemolithoautotrophs, organisms that obtain energy from the oxidation of inorganic substances and carbon from CO2, the one-carbon (C1) metabolism is central to their existence. They have unique pathways to assimilate C1 compounds. Chemolithoautotrophs are microorganisms that derive energy from the oxidation of inorganic compounds and use CO2 as their sole carbon source. Many of these enzymes and pathways are present in chemolithoautotrophic organisms that inhabit hydrothermal vents, where inorganic substances are abundant and can be utilized for energy.
Carbon Monoxide Dehydrogenase/Acetyl-CoA Synthase (CODH/ACS): Relevance to Vent Organisms: Many vent-dwelling bacteria utilize the CODH/ACS complex for carbon fixation by reducing CO2 to CO and synthesizing acetyl-CoA. This pathway is part of the reductive acetyl-CoA pathway, which is used by many thermophilic organisms in hydrothermal vents.
Hydrogenases: Relevance to Vent Organisms: Hydrothermal vent environments are rich in hydrogen, and vent-dwelling microorganisms often use hydrogenases to oxidize hydrogen, generating reducing power for C1 compound reduction.
Formate Dehydrogenase: Relevance to Vent Organisms: Formate dehydrogenase is crucial for many vent-dwelling microorganisms in oxidizing formate to CO2.
Methanogens and Methanotrophs: Relevance to Vent Organisms: Methanogens are common in anaerobic hydrothermal vent environments, where they produce methane from CO2 and other C1 compounds. Methanotrophs in vents can oxidize this methane, converting it back to CO2 or incorporating it into biomass.
Serine Pathway: Some vent-dwelling microorganisms use the serine pathway for C1 assimilation.
Reductive Acetyl-CoA Pathway: This is a significant pathway for CO2 fixation in many thermophilic organisms found in hydrothermal vents.
3-Hydroxypropionate/4-Hydroxybutyrate Cycle: Used by some archaea in hydrothermal vent environments for carbon fixation.
While many of the molecules and enzymes you've listed (SAM, Biotin, Cobalamin, and Folate) are also crucial for one-carbon metabolism in chemolithoautotrophs, these organisms have unique and additional pathways due to their specialized ecological niches and metabolic needs.
[size=16]10.10. Folate Metabolism: A Complex and Essential Cellular Process
The synthesis of folate involves a series of complex enzymatic reactions. Dihydropteroate synthase (DHPS) catalyzes a key step in this pathway, forming 7,8-dihydropteroate from p-aminobenzoate and 6-hydroxymethyl-7,8-dihydropteroate. This reaction links two distinct branches of the folate biosynthesis pathway, demonstrating the interconnected nature of these biochemical processes. Folylpolyglutamate synthase (FPGS) then adds glutamate residues to folates, a critical modification that enhances folate retention within cells and increases their affinity for folate-dependent enzymes. The conversion of dihydrofolate (DHF) to tetrahydrofolate (THF) by dihydrofolate reductase represents another crucial step, maintaining the pool of active folate coenzymes essential for numerous cellular processes.
10.10.1.Folate-Dependent Processes
Folate and its derivatives are integral to several vital cellular functions. In DNA synthesis, folate-dependent enzymes play key roles in the production of purines and thymidylate, essential building blocks of genetic material. Amino acid metabolism heavily relies on folate-mediated one-carbon transfers, particularly in the synthesis of methionine, glycine, and serine. The methylation cycle, crucial for epigenetic regulation and numerous other cellular processes, depends on S-adenosylmethionine (SAM), which is produced through a folate-dependent pathway. These interconnected processes highlight the central role of folate metabolism in maintaining cellular health and function. The intricacy of folate metabolism is evident in the precise structure-function relationships of its enzymes. Each enzyme in the pathway possesses a highly specific active site, tailored to recognize and process particular substrates with remarkable accuracy. This specificity extends to cofactor requirements, reaction mechanisms, and regulatory controls. The interdependence of these enzymes creates a sophisticated network where the product of one reaction becomes the substrate for another, forming a tightly regulated and efficient system. Moreover, the folate cycle demonstrates an impressive level of metabolic plasticity. It can adapt to varying cellular needs, shifting between different one-carbon-carrying forms of folate as required for diverse biochemical reactions. This adaptability is crucial for maintaining cellular homeostasis under varying conditions and metabolic demands.
10.10.2. Utilization of Tetrahydrofolate (THF) Derivatives
Tetrahydrofolate (THF) derivatives are essential for various metabolic processes, including nucleotide synthesis, amino acid metabolism, and methylation reactions. The proper conversion and utilization of these derivatives are crucial for cellular function and growth. The following enzymes play key roles in these transformations:
Methenyltetrahydrofolate cyclohydrolase (EC 3.5.4.9): Smallest known: 182 amino acids (*Aquifex aeolicus*): Catalyzes the conversion of 5,10-methenyltetrahydrofolate to 10-formyltetrahydrofolate. This enzyme is critical for the formation of 10-formyltetrahydrofolate, a key intermediate in purine biosynthesis.
Methylenetetrahydrofolate reductase (EC 1.7.99.5): Smallest known: 187 amino acids (*Thermotoga maritima*): Converts 5,10-methylenetetrahydrofolate to 5-methyltetrahydrofolate. This enzyme is vital for maintaining appropriate levels of 5-methyltetrahydrofolate, which is essential for homocysteine remethylation and methionine synthesis.
Methenyltetrahydrofolate synthetase (EC 6.3.4.3): Smallest known: 222 amino acids (*Aquifex aeolicus*): Converts 5,10-methylenetetrahydrofolate to 5,10-methenyltetrahydrofolate. This enzyme is involved in the interconversion of THF derivatives, facilitating their availability for various metabolic reactions.
5,10-Methenyltetrahydrofolate cyclohydrolase (EC 3.5.4.9): Smallest known: 182 amino acids (*Aquifex aeolicus*): Converts 5,10-methenyltetrahydrofolate to 5,10-methylenetetrahydrofolate. This enzyme is essential for maintaining the balance of methylene and methenyl THF derivatives in the cell.
The THF derivative-related essential enzyme group consists of 4 enzymes. The total number of amino acids for the smallest known versions of these enzymes is 793.
Information on metal clusters or cofactors:
Methenyltetrahydrofolate cyclohydrolase (EC 3.5.4.9): Requires Zn²⁺ as a cofactor for its activity. The zinc ion is crucial for stabilizing the enzyme's structure and facilitating the catalytic reaction.
Methylenetetrahydrofolate reductase (EC 1.7.99.5): Requires FAD as a cofactor for its activity. FAD is essential for the enzyme's function in the reduction of 5,10-methylenetetrahydrofolate to 5-methyltetrahydrofolate.
Methenyltetrahydrofolate synthetase (EC 6.3.4.3): Does not require metal ions or additional cofactors for its catalytic activity.
5,10-Methenyltetrahydrofolate cyclohydrolase (EC 3.5.4.9): Requires Zn²⁺ as a cofactor. The zinc ion aids in the enzyme's catalytic process and stabilization.
10.10.3. Other Related Enzymes in Folate Metabolism
5,10-Methenyltetrahydrofolate cyclohydrolase / 5,10-methylenetetrahydrofolate dehydrogenase.
Glycinamide ribonucleotide formyltransferase (GARFT): Converts glycinamide ribonucleotide (GAR) to formylglycinamide ribonucleotide (FGAR).
10-formyltetrahydrofolate dehydrogenase: Converts 10-formyltetrahydrofolate to CO2, THF, and NADP+.
Methylene tetrahydrofolate dehydrogenase (NADP+).
Challenges in Understanding the Origins of Folate Metabolism
1. Enzyme Complexity and Specificity: Folate metabolism involves highly specialized enzymes with precise structures and functions, raising several questions:
- How did enzymes like dihydropteroate synthase (DHPS) and dihydrofolate reductase (DHFR) acquire their complex structures?
- What mechanisms could account for the development of such high substrate specificity?
- How did these enzymes evolve to catalyze reactions with such remarkable efficiency?
2. Pathway Interdependence: The folate metabolism pathway exhibits a high degree of interdependence among its components, presenting significant challenges:
- How could such an interconnected network of reactions have originated?
- What intermediate forms, if any, could have existed that were functional?
- How did the precise coordination between different enzymes in the pathway develop?
3. Chemical Instability of Folates: The inherent instability of folate molecules presents unique challenges in understanding their role in early biological systems:
- How could these unstable molecules have persisted in early environments?
- What mechanisms could have protected folates from degradation in primitive cells?
- How did the requirement for continuous folate synthesis or intake arise?
4. Dual Nature of Folate Metabolism: The dual role of folate metabolism in one-carbon transfer and redox regulation presents additional challenges:
- How did a single pathway evolve to serve these two distinct cellular functions?
- What mechanisms led to the integration of folate metabolism with cellular redox status?
- How did the complex regulatory systems controlling this dual function originate?
5. Integration with Other Metabolic Pathways: The deep integration of folate metabolism with numerous other cellular processes presents further challenges:
- How did folate metabolism become so linked with other essential pathways?
- What mechanisms coordinated the development of these interconnected systems?
- How can we explain the origin of the complex regulatory mechanisms controlling metabolic flux?
10.11. S-Adenosylmethionine (SAM) Metabolism
S-Adenosylmethionine (SAM) metabolism represents one of the most complex and essential biochemical processes in living organisms. This system, involving numerous enzymes and interconnected pathways, presents significant challenges to our understanding of its origins and development. The synthesis of SAM involves a series of highly specific enzymatic reactions. Methionine adenosyltransferase (MAT) catalyzes the formation of SAM from methionine and ATP. This reaction requires precise molecular recognition and positioning of substrates. The enzyme must overcome significant energetic barriers to form the high-energy sulfonium compound. This molecule contains a positively charged sulfur atom (hence "sulfonium") bonded to three carbon atoms, which makes it energetically unstable and highly reactive, allowing it to readily donate its methyl group in various biochemical reactions.
10.11.1.The SAM-Dependent Methylation Cycle
SAM serves as the primary methyl donor in numerous cellular reactions. Methyltransferases use SAM to methylate DNA, proteins, lipids, and small molecules. This process generates S-adenosylhomocysteine (SAH), which must be efficiently removed to prevent product inhibition. The SAH hydrolase then converts SAH to homocysteine, completing the cycle. The interdependence of these reactions presents a significant challenge to naturalistic explanations. Each step relies on the products of the previous reaction and influences the next, creating a closed loop of metabolic processes. The question arises: how could such a system have emerged gradually when each component depends on the others for functionality?
10.11.2. Regeneration of Methionine
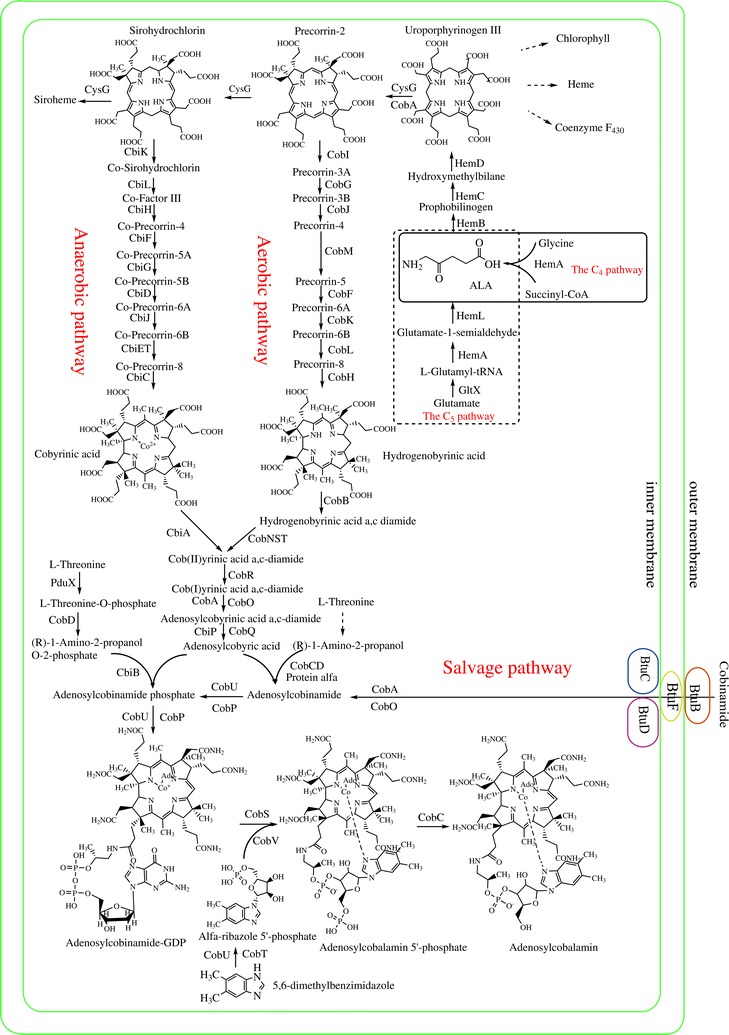
The regeneration of methionine from homocysteine is crucial for maintaining the SAM cycle. This process involves either methionine synthase, which requires vitamin B12 and folate, or betaine-homocysteine methyltransferase. These enzymes exhibit remarkable substrate specificity and catalytic efficiency. The complexity of methionine regeneration, particularly the involvement of cofactors like vitamin B12, adds another layer of complexity to the system. The precise coordination required between these enzymes and their cofactors challenges the notion of a gradual, step-wise development of this metabolic pathway.
10.11.3. Regulation of SAM Metabolism
SAM metabolism is tightly regulated at multiple levels. Allosteric regulation of key enzymes, transcriptional control, and post-translational modifications all play crucial roles in maintaining appropriate SAM levels. This multi-layered regulatory system ensures that SAM concentrations are kept within a narrow range, critical for cellular function. The existence of such sophisticated regulatory mechanisms poses a significant challenge to naturalistic explanations. How could a system with multiple levels of control, each fine-tuned to respond to specific cellular conditions, have arisen through undirected processes?
10.11.4. Integration with Other Metabolic Pathways
SAM metabolism is deeply integrated with numerous other cellular processes, including the folate cycle, transsulfuration pathway, and polyamine synthesis. This network of interdependent reactions raises questions about the origin and development of such interconnected systems. The challenge lies in explaining how these diverse pathways could have become linked without a guiding principle. The precise coordination required between these various metabolic routes suggests a level of complexity that is difficult to account for through random chemical events.
10.11.5. Synthesis of S-Adenosylmethionine (SAM)
S-Adenosylmethionine (SAM) is a vital methyl donor in numerous biological methylation reactions. It is synthesized from methionine and ATP and plays a crucial role in various metabolic processes, including the regulation of gene expression, neurotransmitter synthesis, and lipid metabolism. The pathway for SAM synthesis involves several key enzymes that convert precursors into SAM and other related compounds. Understanding these enzymes and their functions helps elucidate the complexity of SAM metabolism and its biological significance.
Methionine adenosyltransferase (MAT) (EC 2.5.1.6): Smallest known: 228 amino acids (*Escherichia coli*): Catalyzes the conversion of methionine and ATP to S-adenosylmethionine (SAM). This enzyme initiates the SAM synthesis pathway, making it fundamental for the production of this critical methyl donor.
Methylenetetrahydrofolate reductase (MTHFR) (EC 1.5.1.20): Smallest known: 275 amino acids (*Escherichia coli*): Converts 5,10-methylenetetrahydrofolate to 5-methyltetrahydrofolate, which donates a methyl group to homocysteine in the synthesis of methionine. This enzyme is essential for regenerating methionine from homocysteine, indirectly supporting SAM synthesis.
Betaine-homocysteine methyltransferase (BHMT) (EC 2.1.1.5): Smallest known: 360 amino acids (*Escherichia coli*): Utilizes betaine as a methyl donor to convert homocysteine to methionine. This enzyme contributes to the methylation cycle and supports methionine and SAM levels.
Cystathionine β-synthase (CBS) (EC 4.2.1.22): Smallest known: 298 amino acids (*Escherichia coli*): Converts homocysteine to cystathionine as part of the transsulfuration pathway. This enzyme is involved in the metabolism of homocysteine, affecting its availability for SAM synthesis.
The SAM synthesis enzyme group consists of 4 enzymes. The total number of amino acids for the smallest known versions of these enzymes is 1,161.
Information on metal clusters or cofactors:
Methionine adenosyltransferase (MAT) (EC 2.5.1.6): Requires Mg²⁺ as a cofactor for the synthesis of SAM. Magnesium ions are crucial for stabilizing the ATP molecule and facilitating the transfer of the adenosyl group to methionine.
Methylenetetrahydrofolate reductase (MTHFR) (EC 1.5.1.20): Requires FAD (flavin adenine dinucleotide) as a cofactor. FAD is essential for the enzyme's activity in reducing 5,10-methylenetetrahydrofolate to 5-methyltetrahydrofolate.
Betaine-homocysteine methyltransferase (BHMT) (EC 2.1.1.5): Requires Zn²⁺ (zinc ion) as a cofactor. Zinc plays a key role in the enzyme's catalytic activity and stabilization of the active site.
Cystathionine β-synthase (CBS) (EC 4.2.1.22): Requires PLP (pyridoxal phosphate) as a cofactor. PLP is vital for the enzyme's activity in converting homocysteine to cystathionine.
10.11.6. Recycling and Conversion of Tetrahydrofolate (THF)
Tetrahydrofolate (THF) and its derivatives play crucial roles in one-carbon metabolism, which is essential for the synthesis of nucleotides and amino acids. The recycling and conversion of THF are facilitated by several key enzymes, each contributing to the maintenance and utilization of THF derivatives. Here is an overview of the key enzymes involved in this process:
Dihydrofolate reductase (DHFR) (EC 1.5.1.3): Smallest known: 159 amino acids (*Escherichia coli*): Converts dihydrofolate (DHF) to tetrahydrofolate (THF). This enzyme is essential for the regeneration of THF from DHF, ensuring a continuous supply of THF for various metabolic processes.
Serine hydroxymethyltransferase (SHMT) (EC 2.1.2.1): Smallest known: 214 amino acids (*Escherichia coli*): Catalyzes the conversion of serine and THF to glycine and 5,10-methylenetetrahydrofolate. This enzyme is crucial for the transfer of one-carbon units and the production of key THF derivatives involved in nucleotide synthesis.
Folylpolyglutamate synthase (FPGS) (EC 2.5.1.12): Smallest known: 307 amino acids (*Escherichia coli*): Adds glutamate residues to folates to form polyglutamated folates. This enzyme enhances the retention of folates within the cell and increases their effectiveness in metabolic reactions.
Methylenetetrahydrofolate reductase (MTHFR) (EC 1.5.1.20): Smallest known: 275 amino acids (*Escherichia coli*): Converts 5,10-methylenetetrahydrofolate to 5-methyltetrahydrofolate. This enzyme plays a critical role in the methylation cycle, converting THF derivatives to forms needed for methyl group transfer and amino acid metabolism.
Methylene tetrahydrofolate dehydrogenase (MTHFD) (EC 1.5.1.5): Smallest known: 252 amino acids (*Escherichia coli*): Catalyzes the interconversion of various forms of THF. This enzyme is involved in maintaining the balance of THF derivatives required for different metabolic processes.
The THF recycling and conversion enzyme group consists of 5 enzymes. The total number of amino acids for the smallest known versions of these enzymes is 1,447.
Information on metal clusters or cofactors:
Dihydrofolate reductase (DHFR) (EC 1.5.1.3): Requires NADPH as a cofactor for the reduction of dihydrofolate to tetrahydrofolate. NADPH provides the reducing power needed for this reaction.
Serine hydroxymethyltransferase (SHMT) (EC 2.1.2.1): Requires pyridoxal phosphate (PLP) as a cofactor. PLP is crucial for the enzyme's transamination and decarboxylation activities.
Folylpolyglutamate synthase (FPGS) (EC 2.5.1.12): Requires ATP and Mg²⁺ as cofactors. ATP drives the glutamylation reaction, while magnesium ions stabilize the ATP molecule.
Methylenetetrahydrofolate reductase (MTHFR) (EC 1.5.1.20): Requires FAD (flavin adenine dinucleotide) as a cofactor. FAD is essential for the enzyme's reductive activity in the conversion of 5,10-methylenetetrahydrofolate to 5-methyltetrahydrofolate.
Methylene tetrahydrofolate dehydrogenase (MTHFD) (EC 1.5.1.5): Requires NAD⁺ or NADP⁺ as cofactors. These cofactors are necessary for the enzyme's oxidative reactions involving THF derivatives.
10.11.7. Central enzymes and transporters related to the methionine cycle and SAM/SAH metabolism
The methionine cycle and the metabolism of S-adenosylmethionine (SAM) and S-adenosylhomocysteine (SAH) are crucial for cellular methylation processes and the regulation of homocysteine levels. Several key enzymes are involved in these processes:
Methionine adenosyltransferase (MAT) (EC 2.5.1.6): Smallest known: 285 amino acids (*Escherichia coli*): Converts methionine and ATP to S-adenosylmethionine (SAM). This enzyme is central to the methionine cycle, providing SAM, a critical methyl donor for various methylation reactions.
S-adenosylhomocysteine hydrolase (SAHH) (EC 3.3.1.1): Smallest known: 316 amino acids (*Escherichia coli*): Hydrolyzes S-adenosylhomocysteine (SAH) to adenosine and homocysteine. This enzyme is essential for regulating the levels of SAM and SAH, thus controlling methylation reactions and homocysteine metabolism.
Methionine synthase (MS) (EC 2.1.1.13): Smallest known: 755 amino acids (*Bacillus subtilis*): Uses a methyl group from 5-methyltetrahydrofolate to convert homocysteine to methionine. This enzyme is crucial for regenerating methionine, which is essential for maintaining SAM levels and overall methylation balance.
The methionine cycle and SAM/SAH metabolism enzyme group consists of 3 enzymes. The total number of amino acids for the smallest known versions of these enzymes is 1,356.
Information on metal clusters or cofactors:
Methionine adenosyltransferase (MAT) (EC 2.5.1.6): Requires Mg²⁺ as a cofactor. Magnesium ions are essential for the ATP-dependent activation of methionine.
S-adenosylhomocysteine hydrolase (SAHH) (EC 3.3.1.1): Requires Mg²⁺ or Mn²⁺ as cofactors. These metal ions are necessary for the enzyme’s hydrolytic activity on SAH.
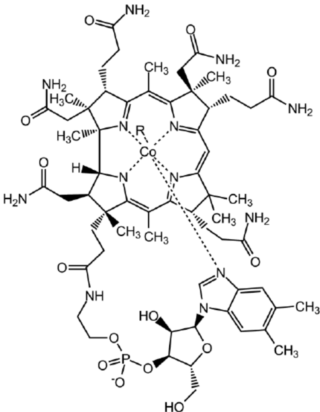
Methionine synthase (MS) (EC 2.1.1.13): Requires vitamin B12 (cobalamin) as a cofactor. Vitamin B12 is essential for the transfer of the methyl group from 5-methyltetrahydrofolate to homocysteine, completing the methionine synthesis.
10.11.8. Methyl transfer with S-adenosylmethionine (SAM)
S-adenosylmethionine (SAM) is a pivotal methyl donor involved in various methylation reactions within the cell, influencing gene expression, protein function, and cellular metabolism. Here is an overview of key components and enzymes involved in methyl transfer with SAM:
S-adenosylmethionine (SAM): Smallest known: Not applicable (SAM is a metabolite rather than a protein): Serves as the principal methyl donor in the cell. SAM provides a methyl group for methylation reactions, which are critical for modifying nucleic acids, proteins, and lipids. The availability of SAM directly affects cellular methylation processes and overall metabolism.
S-adenosylhomocysteine hydrolase (SAHH) (EC 3.3.1.1): Smallest known: 316 amino acids (*Escherichia coli*): Regenerates homocysteine and adenosine from S-adenosylhomocysteine (SAH). This enzyme is essential for maintaining the balance of SAM and SAH, which are crucial for the regulation of methylation reactions and overall cellular methylation status.
The methyl transfer and SAM-related enzyme group consists of 2 components. The total number of amino acids for the smallest known versions of these enzymes is 316 for SAHH. SAM itself is not a protein and does not have an amino acid count.
Information on metal clusters or cofactors:
S-adenosylhomocysteine hydrolase (SAHH) (EC 3.3.1.1): Requires Mg²⁺ or Mn²⁺ as cofactors. These metal ions are essential for the enzyme’s hydrolytic activity on SAH, facilitating the regeneration of homocysteine and adenosine.
This overview highlights the critical role of SAM in methylation and the essential enzyme SAHH in regulating methylation balance by managing SAH levels.
Challenges to Naturalistic Explanations of S-Adenosylmethionine (SAM) Metabolism
1. Complex Enzymatic Reactions and Molecular Recognition: The synthesis and utilization of SAM involve highly specific enzymatic reactions that present significant challenges:
- How did enzymes like methionine adenosyltransferase (MAT) develop the ability to catalyze the formation of the high-energy sulfonium compound SAM?
- What intermediate forms, if any, could have existed that were functional in SAM synthesis?
- How did these enzymes acquire the precise molecular recognition capabilities required for substrate binding and catalysis?
2. Interdependence of Reactions in the SAM-Dependent Methylation Cycle: The cycle of SAM-dependent methylation presents a chicken-and-egg problem:
- How could the cycle have emerged when each step depends on the products of the previous reactions?
- What intermediate forms of this cycle, if any, could have been functional?
- How did the precise coordination between methyltransferases, SAH hydrolase, and methionine regeneration enzymes develop?
3. Cofactor Dependence and Methionine Regeneration: The regeneration of methionine involves complex enzymes and cofactors:
- How did the dependence on vitamin B12 and folate in methionine synthase develop?
- What were the intermediate steps, if any, in the emergence of betaine-homocysteine methyltransferase?
- How did these diverse cofactor requirements become integrated into a single metabolic pathway?
4. Multi-layered Regulation of SAM Metabolism: The tight regulation of SAM metabolism at multiple levels poses significant questions:
- How did such sophisticated regulatory mechanisms develop?
- What intermediate forms of regulation, if any, could have been functional?
- How did the various levels of control (allosteric, transcriptional, post-translational) become integrated?
5. Integration with Other Metabolic Pathways: The deep integration of SAM metabolism with numerous other cellular processes presents challenges:
- How did SAM metabolism become so intricately linked with the folate cycle, transsulfuration pathway, and polyamine synthesis?
- What intermediate stages, if any, could have existed in the development of these interconnections?
- How did the precise coordination required between these pathways arise?
6. Enzyme Specificity and Catalytic Efficiency: The enzymes involved in SAM metabolism display remarkable specificity and efficiency:
- How did these enzymes acquire their high degree of substrate specificity?
- What mechanisms allowed for the development of such catalytic efficiency?
- How do these enzymes maintain their function in the presence of structurally similar molecules?
7. Folate Metabolism and One-Carbon Transfer: The intricate folate metabolism pathway, crucial for SAM synthesis, presents its own set of challenges:
- How did the complex network of enzymes involved in folate metabolism arise?
- What intermediate forms, if any, could have existed in the development of one-carbon transfer reactions?
- How did the precise coordination between folate metabolism and SAM synthesis develop?
8. Compartmentalization and Transport of SAM Metabolites: The cellular compartmentalization of SAM metabolism components poses additional questions:
- How did the specific localization of SAM metabolism enzymes in different cellular compartments develop?
- What mechanisms allowed for the emergence of specific transporters for SAM and related metabolites?
- How did the precise coordination between compartmentalized reactions arise?
These challenges to naturalistic explanations of SAM metabolism highlight the need for further research and careful consideration of alternative hypotheses. The intricate nature of this system, its essential role in cellular function, and the complexity of its components raise significant questions about its origin and development.
10.12. Biotin Biosynthesis
Biotin biosynthesis represents a remarkable feat of biochemical engineering. This process involves a series of highly specific enzymatic reactions, each catalyzed by a unique enzyme with precise substrate recognition capabilities. The pathway begins with pimeloyl-CoA and progresses through several intermediates before culminating in the formation of biotin. The first step in this process involves the condensation of pimeloyl-CoA with L-alanine, catalyzed by 8-amino-7-oxononanoate synthase. This reaction requires precise molecular recognition and positioning of both substrates. The enzyme must overcome significant energetic barriers to form the carbon-nitrogen bond, a process that demands exquisite catalytic prowess. Subsequent steps in the pathway involve equally complex reactions. The conversion of 8-amino-7-oxononanoate to 7,8-diaminononanoate, catalyzed by 8-amino-7-oxononanoate aminotransferase, requires the transfer of an amino group from a donor molecule. This reaction demands not only substrate specificity but also the ability to facilitate the transfer of chemical groups between molecules. The formation of dethiobiotin from 7,8-diaminononanoate, catalyzed by dethiobiotin synthetase, involves the ATP-dependent closure of a ureido ring. This step represents a significant increase in molecular complexity, requiring precise control over the reaction trajectory to ensure the correct product is formed. The final step, the conversion of dethiobiotin to biotin by biotin synthase, is perhaps the most remarkable. This reaction involves the insertion of a sulfur atom into an unactivated carbon-hydrogen bond, a feat that pushes the boundaries of known biochemistry. The enzyme employs a complex iron-sulfur cluster and S-adenosyl methionine as a radical initiator to accomplish this challenging transformation.
10.12.1. Enzyme Specificity and Catalytic Efficiency
The enzymes involved in biotin biosynthesis exhibit remarkable substrate specificity and catalytic efficiency. Each enzyme in the pathway must recognize its specific substrate among a sea of structurally similar molecules within the cell. This level of discrimination requires precisely shaped binding pockets and intricate networks of chemical interactions between the enzyme and its substrate. Moreover, these enzymes catalyze their respective reactions with extraordinary efficiency. They accelerate reaction rates by factors of millions or even billions, allowing the cell to produce biotin on biologically relevant timescales. This catalytic prowess is achieved through complex mechanisms involving precisely positioned catalytic residues, controlled micro-environments within the active site, and dynamic conformational changes during the catalytic cycle.
10.12.2. Pathway Integration and Regulation
The biotin biosynthesis pathway does not exist in isolation but is intimately connected with other cellular processes. The pathway's starting material, pimeloyl-CoA, intersects with fatty acid metabolism. The pathway also connects with amino acid metabolism through the use of L-alanine and the aminotransferase reaction. Furthermore, the final step requires S-adenosyl methionine, linking biotin synthesis to one-carbon metabolism. This integration demands precise regulation to ensure that biotin production matches cellular needs without depleting resources required for other essential processes. The pathway is subject to complex regulatory mechanisms, including feedback inhibition and transcriptional control. These regulatory systems must have developed in concert with the biosynthetic pathway itself, adding another layer of complexity to the system.
10.12.3. Cofactor Dependence
Several steps in the biotin biosynthesis pathway require specific cofactors. The aminotransferase reaction depends on pyridoxal phosphate, while the final sulfur insertion step requires both an iron-sulfur cluster and S-adenosyl methionine. The dependence on these cofactors raises additional questions about the origin of the pathway. How did these enzymes develop their ability to bind and utilize these complex cofactors? How did the cell ensure the availability of these cofactors in concert with the development of the biotin synthesis pathway?
Biotin biosynthesis is a crucial metabolic pathway that produces biotin (vitamin B7), an essential cofactor for carboxylase enzymes involved in fatty acid synthesis, gluconeogenesis, and amino acid metabolism. This pathway is present in many bacteria, fungi, and plants, but most animals, including humans, lack the ability to synthesize biotin and must obtain it from their diet. The biotin biosynthesis pathway is of significant interest due to its potential as a target for antimicrobial drugs and its importance in industrial biotechnology.
Key enzymes involved:
Lysine 6-aminotransferase (EC 2.6.1.36): Smallest known: 405 amino acids (Thermus thermophilus). This enzyme catalyzes the first step in biotin biosynthesis, converting L-lysine to L-2,6-diaminopimelate. It plays a crucial role in initiating the pathway and is essential for organisms that synthesize biotin de novo.
7,8-Diaminononanoate synthase (EC 6.3.1.25): Smallest known: 384 amino acids (Aquifex aeolicus). This enzyme catalyzes the synthesis of 7,8-diaminononanoate from 7-keto-8-aminopelargonic acid and S-adenosyl methionine. It is critical for the formation of the carbon skeleton of biotin.
Dethiobiotin synthetase (EC 6.3.3.3): Smallest known: 224 amino acids (Helicobacter pylori). This enzyme catalyzes the formation of dethiobiotin from 7,8-diaminononanoate. It is essential for creating the ureido ring structure characteristic of biotin.
Biotin synthase (EC 2.8.1.6): Smallest known: 316 amino acids (Bacillus subtilis). This enzyme catalyzes the final step in biotin biosynthesis, converting dethiobiotin to biotin. It is crucial for completing the biotin structure and is often considered the rate-limiting step in the pathway.
The biotin biosynthesis essential enzyme group consists of 4 enzymes. The total number of amino acids for the smallest known versions of these enzymes is 1,329.
Information on metal clusters or cofactors:
Lysine 6-aminotransferase (EC 2.6.1.36): Requires pyridoxal 5'-phosphate (PLP) as a cofactor. PLP is covalently bound to a lysine residue in the active site and is essential for the transamination reaction.
7,8-Diaminononanoate synthase (EC 6.3.1.25): Requires ATP and Mg²⁺ for its catalytic activity. The magnesium ion acts as a cofactor, facilitating the ATP-dependent reaction.
Dethiobiotin synthetase (EC 6.3.3.3): Requires ATP and Mg²⁺ for its catalytic activity. The magnesium ion is essential for ATP binding and the subsequent reaction.
Biotin synthase (EC 2.8.1.6): Contains an iron-sulfur cluster ([4Fe-4S]) and requires S-adenosyl methionine (SAM) as a cofactor. The iron-sulfur cluster is crucial for the radical SAM mechanism used to insert the sulfur atom into dethiobiotin.
This overview highlights the complexity and importance of the biotin biosynthesis pathway, emphasizing the unique roles of each enzyme and their cofactor requirements. The pathway's absence in most animals makes it an attractive target for antimicrobial drug development, while its presence in certain microorganisms is leveraged for industrial biotin production.
Utilization of Biotin
Acetyl-CoA carboxylase: EC: 6.4.1.2 - Utilizes biotin to carboxylate acetyl-CoA to malonyl-CoA.
Recycling and Conversion of Biotin
Biotinidase: EC: 3.5.1.76 Hydrolyzes biocytin to release biotin for recycling.
Biotinidase: EC: 3.5.1.76 - Hydrolyzes biocytin to release biotin for recycling.
Challenges in Understanding Biotinidase Function and Regulation
Biotinidase exhibits remarkable specificity in recognizing and hydrolyzing biocytin. This presents several challenging questions:
- How did biotinidase develop its precise active site configuration to accommodate biocytin?
- What intermediate forms, if any, could have existed that were functional in biotin recycling?
- How does biotinidase distinguish biocytin from structurally similar molecules in the cellular milieu?
1. Catalytic Mechanism and Efficiency: The catalytic mechanism of biotinidase involves complex proton transfers and nucleophilic attack. This raises several questions:
- How did the precise arrangement of catalytic residues in biotinidase's active site arise?
- What is the exact sequence of chemical events during catalysis, and how is it coordinated?
- How does biotinidase achieve its high catalytic efficiency?
2. Regulation of Biotinidase Activity: The regulation of biotinidase activity is crucial for maintaining proper biotin levels. This presents several challenges:
- How is biotinidase activity coordinated with biotin synthesis and utilization?
- What mechanisms control biotinidase expression and activity in response to cellular biotin levels?
- How did these regulatory mechanisms develop in concert with biotinidase itself?
Studies by Pindolia et al. (2011) have shown that biotinidase expression is regulated by biotin status, but the molecular details of this regulation remain unclear.
4. Multifunctionality of Biotinidase: Biotinidase has been found to have functions beyond biotin recycling, including a potential role in processing biotinylated histones. This raises several questions:
- How did biotinidase acquire these additional functions?
- What is the relationship between biotinidase's different functions?
- How does the cell regulate these diverse activities?
5. Biotinidase Deficiency and Genetic Variations: Biotinidase deficiency is a genetic disorder with varying degrees of severity. This presents several challenges:
- How do different mutations in the biotinidase gene affect enzyme function?
- What is the relationship between enzyme structure and the various clinical presentations of biotinidase deficiency?
- How has biotinidase maintained its function despite genetic variations across populations?
The study of biotinidase presents numerous challenges that defy simple explanations. The enzyme's structural complexity, catalytic sophistication, regulatory mechanisms, and multifunctionality raise profound questions about its origins and development. Current research continues to uncover new aspects of biotinidase function, but many fundamental questions remain unanswered. Understanding these aspects fully will require interdisciplinary approaches and novel experimental techniques.
10.13. Carbon Monoxide Dehydrogenase (CODH): A Marvel of Biochemical Engineering
Carbon Monoxide Dehydrogenase (CODH) represents a remarkable feat of biochemical engineering, playing a crucial role in carbon cycling and autotrophic growth in certain microorganisms.
CODH is named for its primary function:
1. "Carbon Monoxide" refers to its substrate, CO.
2. "Dehydrogenase" indicates its role in removing hydrogen (in this case, as part of oxidizing CO to CO2).
The name reflects the enzyme's ability to catalyze the oxidation of carbon monoxide (CO) to carbon dioxide (CO2), effectively removing hydrogen from the substrate. While some CODHs can also catalyze the reverse reaction, the name emphasizes its historically first-discovered and most prominent function. This enzyme catalyzes the interconversion of carbon monoxide (CO) and carbon dioxide (CO2), a reaction central to the Wood-Ljungdahl carbon fixation pathway. The complexity and efficiency of CODH raises questions about its origin and function, challenging our understanding of biochemical systems. CODH exists in two main forms: the monofunctional CODH (EC: 1.2.99.2) and the bifunctional CODH/Acetyl-CoA Synthase (CODH/ACS) complex (EC: 1.2.7.4). The monofunctional CODH primarily oxidizes CO to CO2, while the bifunctional complex not only catalyzes this reaction but also participates in the synthesis of acetyl-CoA from CO2, CO, and a methyl group. These enzymes demonstrate extraordinary catalytic prowess, operating at the thermodynamic limit with minimal overpotential.
10.13.1. Catalytic Efficiency
CODHs are among the most efficient enzymes known, operating near the thermodynamic limit of the CO/CO2 interconversion reaction. This means they catalyze the reaction with minimal energy loss, achieving maximum possible efficiency. The turnover rates (kcat) of CODHs are impressively high:
- For CO oxidation: up to 40,000 s⁻¹
- For CO2 reduction: up to 12 s⁻¹
These rates are among the highest observed for metalloenzymes, especially considering the complexity of the reaction. CODHs operate with an overpotential of only about 90 mV for CO oxidation. This is remarkably low, especially when compared to synthetic catalysts which typically require overpotentials of 400-600 mV for similar reactions. Imagine you're trying to push a heavy box up a small hill. The hill represents the energy barrier that needs to be overcome for a chemical reaction to occur. In an ideal world, you'd only need to exert exactly enough energy to get the box to the top of the hill. This "ideal" amount of energy is like the theoretical minimum energy needed for a chemical reaction. Now, in reality, you might need to push a bit harder than this ideal minimum to get the box moving and overcome friction. This extra push is similar to what we call "overpotential" in chemistry. In the context of Carbon Monoxide Dehydrogenase (CODH) enzymes:
The "hill" is the energy barrier for converting CO to CO2.
The "ideal push" is the theoretical minimum energy (or voltage) needed to make this conversion happen.
The "extra push" (overpotential) is the additional energy the enzyme actually needs to use above this theoretical minimum.
When we say CODHs operate with an overpotential of only about 90 mV for CO oxidation, it means:
- These enzymes need only a tiny bit of extra energy (90 millivolts) beyond the theoretical minimum to catalyze the reaction.
- This is remarkably low - like needing only a small extra push to get that heavy box over the hill.
- Most artificial catalysts we've created for similar reactions need a much bigger "extra push" - often 4-6 times more (400-600 mV).
To put it in everyday terms, it's like CODH enzymes are incredibly efficient cars that can climb a hill using just a touch more gas than the absolute minimum required. In contrast, many of our artificial catalysts are like less efficient vehicles that need to rev their engines much harder to climb the same hill. This extremely low overpotential is one of the reasons why CODHs are considered so remarkably efficient. They're doing a complex chemical job with very little wasted energy.
10.13.2. Mechanisms of Efficiency
1. Optimized Active Site Structure: The [NiFe4S4] C-cluster is precisely arranged to facilitate electron transfer and substrate binding. The asymmetric position of the nickel ion allows for optimal interaction with CO and CO2. The [NiFe4S4] C-cluster at the heart of Carbon Monoxide Dehydrogenase (CODH) is indeed a marvel of biochemical engineering, showcasing an extraordinary level of complexity and precision. Let's break down the intricacies of this structure and its assembly:

a Crystal structure of ACS/CODH complex (Darnault et al. 2003). ACS forms the bifunctional enzyme with carbon monoxide dehydrogenase (CODH), which converts carbon dioxide into carbon monoxide, AcetylCoA Synthase/Carbon Monoxide Dehydrogenase (ACS/CODH). The structure of the CODH/ACS enzyme consists of the CODH enzyme as a dimer at the center with two ACS subunits on each side (Ragsdale 2004). b Structure of A-cluster (Svetlitchnyi et al. 2004). c Structure of C-cluster (Dobbek et al. 2001) 1
[/size]
Chemolithoautotrophs, which obtain energy from inorganic substances and carbon from CO2, are studied separately as potential models for early life forms due to their unique metabolic pathways and ability to thrive in extreme conditions. Their specialized enzymes, such as carbon monoxide dehydrogenase/acetyl-CoA synthase and hydrogenases, offer insights into primitive biochemical processes that could have existed in early Earth environments like hydrothermal vents. The study of one-carbon metabolism in both widespread and chemolithoautotrophic pathways provides a comprehensive approach to understanding the possible origins of life. In chemolithoautotrophs, organisms that obtain energy from the oxidation of inorganic substances and carbon from CO2, the one-carbon (C1) metabolism is central to their existence. They have unique pathways to assimilate C1 compounds. Chemolithoautotrophs are microorganisms that derive energy from the oxidation of inorganic compounds and use CO2 as their sole carbon source. Many of these enzymes and pathways are present in chemolithoautotrophic organisms that inhabit hydrothermal vents, where inorganic substances are abundant and can be utilized for energy.
Carbon Monoxide Dehydrogenase/Acetyl-CoA Synthase (CODH/ACS): Relevance to Vent Organisms: Many vent-dwelling bacteria utilize the CODH/ACS complex for carbon fixation by reducing CO2 to CO and synthesizing acetyl-CoA. This pathway is part of the reductive acetyl-CoA pathway, which is used by many thermophilic organisms in hydrothermal vents.
Hydrogenases: Relevance to Vent Organisms: Hydrothermal vent environments are rich in hydrogen, and vent-dwelling microorganisms often use hydrogenases to oxidize hydrogen, generating reducing power for C1 compound reduction.
Formate Dehydrogenase: Relevance to Vent Organisms: Formate dehydrogenase is crucial for many vent-dwelling microorganisms in oxidizing formate to CO2.
Methanogens and Methanotrophs: Relevance to Vent Organisms: Methanogens are common in anaerobic hydrothermal vent environments, where they produce methane from CO2 and other C1 compounds. Methanotrophs in vents can oxidize this methane, converting it back to CO2 or incorporating it into biomass.
Serine Pathway: Some vent-dwelling microorganisms use the serine pathway for C1 assimilation.
Reductive Acetyl-CoA Pathway: This is a significant pathway for CO2 fixation in many thermophilic organisms found in hydrothermal vents.
3-Hydroxypropionate/4-Hydroxybutyrate Cycle: Used by some archaea in hydrothermal vent environments for carbon fixation.
While many of the molecules and enzymes you've listed (SAM, Biotin, Cobalamin, and Folate) are also crucial for one-carbon metabolism in chemolithoautotrophs, these organisms have unique and additional pathways due to their specialized ecological niches and metabolic needs.
[size=16]10.10. Folate Metabolism: A Complex and Essential Cellular Process
The synthesis of folate involves a series of complex enzymatic reactions. Dihydropteroate synthase (DHPS) catalyzes a key step in this pathway, forming 7,8-dihydropteroate from p-aminobenzoate and 6-hydroxymethyl-7,8-dihydropteroate. This reaction links two distinct branches of the folate biosynthesis pathway, demonstrating the interconnected nature of these biochemical processes. Folylpolyglutamate synthase (FPGS) then adds glutamate residues to folates, a critical modification that enhances folate retention within cells and increases their affinity for folate-dependent enzymes. The conversion of dihydrofolate (DHF) to tetrahydrofolate (THF) by dihydrofolate reductase represents another crucial step, maintaining the pool of active folate coenzymes essential for numerous cellular processes.
10.10.1.Folate-Dependent Processes
Folate and its derivatives are integral to several vital cellular functions. In DNA synthesis, folate-dependent enzymes play key roles in the production of purines and thymidylate, essential building blocks of genetic material. Amino acid metabolism heavily relies on folate-mediated one-carbon transfers, particularly in the synthesis of methionine, glycine, and serine. The methylation cycle, crucial for epigenetic regulation and numerous other cellular processes, depends on S-adenosylmethionine (SAM), which is produced through a folate-dependent pathway. These interconnected processes highlight the central role of folate metabolism in maintaining cellular health and function. The intricacy of folate metabolism is evident in the precise structure-function relationships of its enzymes. Each enzyme in the pathway possesses a highly specific active site, tailored to recognize and process particular substrates with remarkable accuracy. This specificity extends to cofactor requirements, reaction mechanisms, and regulatory controls. The interdependence of these enzymes creates a sophisticated network where the product of one reaction becomes the substrate for another, forming a tightly regulated and efficient system. Moreover, the folate cycle demonstrates an impressive level of metabolic plasticity. It can adapt to varying cellular needs, shifting between different one-carbon-carrying forms of folate as required for diverse biochemical reactions. This adaptability is crucial for maintaining cellular homeostasis under varying conditions and metabolic demands.
10.10.2. Utilization of Tetrahydrofolate (THF) Derivatives
Tetrahydrofolate (THF) derivatives are essential for various metabolic processes, including nucleotide synthesis, amino acid metabolism, and methylation reactions. The proper conversion and utilization of these derivatives are crucial for cellular function and growth. The following enzymes play key roles in these transformations:
Methenyltetrahydrofolate cyclohydrolase (EC 3.5.4.9): Smallest known: 182 amino acids (*Aquifex aeolicus*): Catalyzes the conversion of 5,10-methenyltetrahydrofolate to 10-formyltetrahydrofolate. This enzyme is critical for the formation of 10-formyltetrahydrofolate, a key intermediate in purine biosynthesis.
Methylenetetrahydrofolate reductase (EC 1.7.99.5): Smallest known: 187 amino acids (*Thermotoga maritima*): Converts 5,10-methylenetetrahydrofolate to 5-methyltetrahydrofolate. This enzyme is vital for maintaining appropriate levels of 5-methyltetrahydrofolate, which is essential for homocysteine remethylation and methionine synthesis.
Methenyltetrahydrofolate synthetase (EC 6.3.4.3): Smallest known: 222 amino acids (*Aquifex aeolicus*): Converts 5,10-methylenetetrahydrofolate to 5,10-methenyltetrahydrofolate. This enzyme is involved in the interconversion of THF derivatives, facilitating their availability for various metabolic reactions.
5,10-Methenyltetrahydrofolate cyclohydrolase (EC 3.5.4.9): Smallest known: 182 amino acids (*Aquifex aeolicus*): Converts 5,10-methenyltetrahydrofolate to 5,10-methylenetetrahydrofolate. This enzyme is essential for maintaining the balance of methylene and methenyl THF derivatives in the cell.
The THF derivative-related essential enzyme group consists of 4 enzymes. The total number of amino acids for the smallest known versions of these enzymes is 793.
Information on metal clusters or cofactors:
Methenyltetrahydrofolate cyclohydrolase (EC 3.5.4.9): Requires Zn²⁺ as a cofactor for its activity. The zinc ion is crucial for stabilizing the enzyme's structure and facilitating the catalytic reaction.
Methylenetetrahydrofolate reductase (EC 1.7.99.5): Requires FAD as a cofactor for its activity. FAD is essential for the enzyme's function in the reduction of 5,10-methylenetetrahydrofolate to 5-methyltetrahydrofolate.
Methenyltetrahydrofolate synthetase (EC 6.3.4.3): Does not require metal ions or additional cofactors for its catalytic activity.
5,10-Methenyltetrahydrofolate cyclohydrolase (EC 3.5.4.9): Requires Zn²⁺ as a cofactor. The zinc ion aids in the enzyme's catalytic process and stabilization.
10.10.3. Other Related Enzymes in Folate Metabolism
5,10-Methenyltetrahydrofolate cyclohydrolase / 5,10-methylenetetrahydrofolate dehydrogenase.
Glycinamide ribonucleotide formyltransferase (GARFT): Converts glycinamide ribonucleotide (GAR) to formylglycinamide ribonucleotide (FGAR).
10-formyltetrahydrofolate dehydrogenase: Converts 10-formyltetrahydrofolate to CO2, THF, and NADP+.
Methylene tetrahydrofolate dehydrogenase (NADP+).
Challenges in Understanding the Origins of Folate Metabolism
1. Enzyme Complexity and Specificity: Folate metabolism involves highly specialized enzymes with precise structures and functions, raising several questions:
- How did enzymes like dihydropteroate synthase (DHPS) and dihydrofolate reductase (DHFR) acquire their complex structures?
- What mechanisms could account for the development of such high substrate specificity?
- How did these enzymes evolve to catalyze reactions with such remarkable efficiency?
2. Pathway Interdependence: The folate metabolism pathway exhibits a high degree of interdependence among its components, presenting significant challenges:
- How could such an interconnected network of reactions have originated?
- What intermediate forms, if any, could have existed that were functional?
- How did the precise coordination between different enzymes in the pathway develop?
3. Chemical Instability of Folates: The inherent instability of folate molecules presents unique challenges in understanding their role in early biological systems:
- How could these unstable molecules have persisted in early environments?
- What mechanisms could have protected folates from degradation in primitive cells?
- How did the requirement for continuous folate synthesis or intake arise?
4. Dual Nature of Folate Metabolism: The dual role of folate metabolism in one-carbon transfer and redox regulation presents additional challenges:
- How did a single pathway evolve to serve these two distinct cellular functions?
- What mechanisms led to the integration of folate metabolism with cellular redox status?
- How did the complex regulatory systems controlling this dual function originate?
5. Integration with Other Metabolic Pathways: The deep integration of folate metabolism with numerous other cellular processes presents further challenges:
- How did folate metabolism become so linked with other essential pathways?
- What mechanisms coordinated the development of these interconnected systems?
- How can we explain the origin of the complex regulatory mechanisms controlling metabolic flux?
10.11. S-Adenosylmethionine (SAM) Metabolism
S-Adenosylmethionine (SAM) metabolism represents one of the most complex and essential biochemical processes in living organisms. This system, involving numerous enzymes and interconnected pathways, presents significant challenges to our understanding of its origins and development. The synthesis of SAM involves a series of highly specific enzymatic reactions. Methionine adenosyltransferase (MAT) catalyzes the formation of SAM from methionine and ATP. This reaction requires precise molecular recognition and positioning of substrates. The enzyme must overcome significant energetic barriers to form the high-energy sulfonium compound. This molecule contains a positively charged sulfur atom (hence "sulfonium") bonded to three carbon atoms, which makes it energetically unstable and highly reactive, allowing it to readily donate its methyl group in various biochemical reactions.
10.11.1.The SAM-Dependent Methylation Cycle
SAM serves as the primary methyl donor in numerous cellular reactions. Methyltransferases use SAM to methylate DNA, proteins, lipids, and small molecules. This process generates S-adenosylhomocysteine (SAH), which must be efficiently removed to prevent product inhibition. The SAH hydrolase then converts SAH to homocysteine, completing the cycle. The interdependence of these reactions presents a significant challenge to naturalistic explanations. Each step relies on the products of the previous reaction and influences the next, creating a closed loop of metabolic processes. The question arises: how could such a system have emerged gradually when each component depends on the others for functionality?
10.11.2. Regeneration of Methionine
The regeneration of methionine from homocysteine is crucial for maintaining the SAM cycle. This process involves either methionine synthase, which requires vitamin B12 and folate, or betaine-homocysteine methyltransferase. These enzymes exhibit remarkable substrate specificity and catalytic efficiency. The complexity of methionine regeneration, particularly the involvement of cofactors like vitamin B12, adds another layer of complexity to the system. The precise coordination required between these enzymes and their cofactors challenges the notion of a gradual, step-wise development of this metabolic pathway.
10.11.3. Regulation of SAM Metabolism
SAM metabolism is tightly regulated at multiple levels. Allosteric regulation of key enzymes, transcriptional control, and post-translational modifications all play crucial roles in maintaining appropriate SAM levels. This multi-layered regulatory system ensures that SAM concentrations are kept within a narrow range, critical for cellular function. The existence of such sophisticated regulatory mechanisms poses a significant challenge to naturalistic explanations. How could a system with multiple levels of control, each fine-tuned to respond to specific cellular conditions, have arisen through undirected processes?
10.11.4. Integration with Other Metabolic Pathways
SAM metabolism is deeply integrated with numerous other cellular processes, including the folate cycle, transsulfuration pathway, and polyamine synthesis. This network of interdependent reactions raises questions about the origin and development of such interconnected systems. The challenge lies in explaining how these diverse pathways could have become linked without a guiding principle. The precise coordination required between these various metabolic routes suggests a level of complexity that is difficult to account for through random chemical events.
10.11.5. Synthesis of S-Adenosylmethionine (SAM)
S-Adenosylmethionine (SAM) is a vital methyl donor in numerous biological methylation reactions. It is synthesized from methionine and ATP and plays a crucial role in various metabolic processes, including the regulation of gene expression, neurotransmitter synthesis, and lipid metabolism. The pathway for SAM synthesis involves several key enzymes that convert precursors into SAM and other related compounds. Understanding these enzymes and their functions helps elucidate the complexity of SAM metabolism and its biological significance.
Methionine adenosyltransferase (MAT) (EC 2.5.1.6): Smallest known: 228 amino acids (*Escherichia coli*): Catalyzes the conversion of methionine and ATP to S-adenosylmethionine (SAM). This enzyme initiates the SAM synthesis pathway, making it fundamental for the production of this critical methyl donor.
Methylenetetrahydrofolate reductase (MTHFR) (EC 1.5.1.20): Smallest known: 275 amino acids (*Escherichia coli*): Converts 5,10-methylenetetrahydrofolate to 5-methyltetrahydrofolate, which donates a methyl group to homocysteine in the synthesis of methionine. This enzyme is essential for regenerating methionine from homocysteine, indirectly supporting SAM synthesis.
Betaine-homocysteine methyltransferase (BHMT) (EC 2.1.1.5): Smallest known: 360 amino acids (*Escherichia coli*): Utilizes betaine as a methyl donor to convert homocysteine to methionine. This enzyme contributes to the methylation cycle and supports methionine and SAM levels.
Cystathionine β-synthase (CBS) (EC 4.2.1.22): Smallest known: 298 amino acids (*Escherichia coli*): Converts homocysteine to cystathionine as part of the transsulfuration pathway. This enzyme is involved in the metabolism of homocysteine, affecting its availability for SAM synthesis.
The SAM synthesis enzyme group consists of 4 enzymes. The total number of amino acids for the smallest known versions of these enzymes is 1,161.
Information on metal clusters or cofactors:
Methionine adenosyltransferase (MAT) (EC 2.5.1.6): Requires Mg²⁺ as a cofactor for the synthesis of SAM. Magnesium ions are crucial for stabilizing the ATP molecule and facilitating the transfer of the adenosyl group to methionine.
Methylenetetrahydrofolate reductase (MTHFR) (EC 1.5.1.20): Requires FAD (flavin adenine dinucleotide) as a cofactor. FAD is essential for the enzyme's activity in reducing 5,10-methylenetetrahydrofolate to 5-methyltetrahydrofolate.
Betaine-homocysteine methyltransferase (BHMT) (EC 2.1.1.5): Requires Zn²⁺ (zinc ion) as a cofactor. Zinc plays a key role in the enzyme's catalytic activity and stabilization of the active site.
Cystathionine β-synthase (CBS) (EC 4.2.1.22): Requires PLP (pyridoxal phosphate) as a cofactor. PLP is vital for the enzyme's activity in converting homocysteine to cystathionine.
10.11.6. Recycling and Conversion of Tetrahydrofolate (THF)
Tetrahydrofolate (THF) and its derivatives play crucial roles in one-carbon metabolism, which is essential for the synthesis of nucleotides and amino acids. The recycling and conversion of THF are facilitated by several key enzymes, each contributing to the maintenance and utilization of THF derivatives. Here is an overview of the key enzymes involved in this process:
Dihydrofolate reductase (DHFR) (EC 1.5.1.3): Smallest known: 159 amino acids (*Escherichia coli*): Converts dihydrofolate (DHF) to tetrahydrofolate (THF). This enzyme is essential for the regeneration of THF from DHF, ensuring a continuous supply of THF for various metabolic processes.
Serine hydroxymethyltransferase (SHMT) (EC 2.1.2.1): Smallest known: 214 amino acids (*Escherichia coli*): Catalyzes the conversion of serine and THF to glycine and 5,10-methylenetetrahydrofolate. This enzyme is crucial for the transfer of one-carbon units and the production of key THF derivatives involved in nucleotide synthesis.
Folylpolyglutamate synthase (FPGS) (EC 2.5.1.12): Smallest known: 307 amino acids (*Escherichia coli*): Adds glutamate residues to folates to form polyglutamated folates. This enzyme enhances the retention of folates within the cell and increases their effectiveness in metabolic reactions.
Methylenetetrahydrofolate reductase (MTHFR) (EC 1.5.1.20): Smallest known: 275 amino acids (*Escherichia coli*): Converts 5,10-methylenetetrahydrofolate to 5-methyltetrahydrofolate. This enzyme plays a critical role in the methylation cycle, converting THF derivatives to forms needed for methyl group transfer and amino acid metabolism.
Methylene tetrahydrofolate dehydrogenase (MTHFD) (EC 1.5.1.5): Smallest known: 252 amino acids (*Escherichia coli*): Catalyzes the interconversion of various forms of THF. This enzyme is involved in maintaining the balance of THF derivatives required for different metabolic processes.
The THF recycling and conversion enzyme group consists of 5 enzymes. The total number of amino acids for the smallest known versions of these enzymes is 1,447.
Information on metal clusters or cofactors:
Dihydrofolate reductase (DHFR) (EC 1.5.1.3): Requires NADPH as a cofactor for the reduction of dihydrofolate to tetrahydrofolate. NADPH provides the reducing power needed for this reaction.
Serine hydroxymethyltransferase (SHMT) (EC 2.1.2.1): Requires pyridoxal phosphate (PLP) as a cofactor. PLP is crucial for the enzyme's transamination and decarboxylation activities.
Folylpolyglutamate synthase (FPGS) (EC 2.5.1.12): Requires ATP and Mg²⁺ as cofactors. ATP drives the glutamylation reaction, while magnesium ions stabilize the ATP molecule.
Methylenetetrahydrofolate reductase (MTHFR) (EC 1.5.1.20): Requires FAD (flavin adenine dinucleotide) as a cofactor. FAD is essential for the enzyme's reductive activity in the conversion of 5,10-methylenetetrahydrofolate to 5-methyltetrahydrofolate.
Methylene tetrahydrofolate dehydrogenase (MTHFD) (EC 1.5.1.5): Requires NAD⁺ or NADP⁺ as cofactors. These cofactors are necessary for the enzyme's oxidative reactions involving THF derivatives.
10.11.7. Central enzymes and transporters related to the methionine cycle and SAM/SAH metabolism
The methionine cycle and the metabolism of S-adenosylmethionine (SAM) and S-adenosylhomocysteine (SAH) are crucial for cellular methylation processes and the regulation of homocysteine levels. Several key enzymes are involved in these processes:
Methionine adenosyltransferase (MAT) (EC 2.5.1.6): Smallest known: 285 amino acids (*Escherichia coli*): Converts methionine and ATP to S-adenosylmethionine (SAM). This enzyme is central to the methionine cycle, providing SAM, a critical methyl donor for various methylation reactions.
S-adenosylhomocysteine hydrolase (SAHH) (EC 3.3.1.1): Smallest known: 316 amino acids (*Escherichia coli*): Hydrolyzes S-adenosylhomocysteine (SAH) to adenosine and homocysteine. This enzyme is essential for regulating the levels of SAM and SAH, thus controlling methylation reactions and homocysteine metabolism.
Methionine synthase (MS) (EC 2.1.1.13): Smallest known: 755 amino acids (*Bacillus subtilis*): Uses a methyl group from 5-methyltetrahydrofolate to convert homocysteine to methionine. This enzyme is crucial for regenerating methionine, which is essential for maintaining SAM levels and overall methylation balance.
The methionine cycle and SAM/SAH metabolism enzyme group consists of 3 enzymes. The total number of amino acids for the smallest known versions of these enzymes is 1,356.
Information on metal clusters or cofactors:
Methionine adenosyltransferase (MAT) (EC 2.5.1.6): Requires Mg²⁺ as a cofactor. Magnesium ions are essential for the ATP-dependent activation of methionine.
S-adenosylhomocysteine hydrolase (SAHH) (EC 3.3.1.1): Requires Mg²⁺ or Mn²⁺ as cofactors. These metal ions are necessary for the enzyme’s hydrolytic activity on SAH.
Methionine synthase (MS) (EC 2.1.1.13): Requires vitamin B12 (cobalamin) as a cofactor. Vitamin B12 is essential for the transfer of the methyl group from 5-methyltetrahydrofolate to homocysteine, completing the methionine synthesis.
10.11.8. Methyl transfer with S-adenosylmethionine (SAM)
S-adenosylmethionine (SAM) is a pivotal methyl donor involved in various methylation reactions within the cell, influencing gene expression, protein function, and cellular metabolism. Here is an overview of key components and enzymes involved in methyl transfer with SAM:
S-adenosylmethionine (SAM): Smallest known: Not applicable (SAM is a metabolite rather than a protein): Serves as the principal methyl donor in the cell. SAM provides a methyl group for methylation reactions, which are critical for modifying nucleic acids, proteins, and lipids. The availability of SAM directly affects cellular methylation processes and overall metabolism.
S-adenosylhomocysteine hydrolase (SAHH) (EC 3.3.1.1): Smallest known: 316 amino acids (*Escherichia coli*): Regenerates homocysteine and adenosine from S-adenosylhomocysteine (SAH). This enzyme is essential for maintaining the balance of SAM and SAH, which are crucial for the regulation of methylation reactions and overall cellular methylation status.
The methyl transfer and SAM-related enzyme group consists of 2 components. The total number of amino acids for the smallest known versions of these enzymes is 316 for SAHH. SAM itself is not a protein and does not have an amino acid count.
Information on metal clusters or cofactors:
S-adenosylhomocysteine hydrolase (SAHH) (EC 3.3.1.1): Requires Mg²⁺ or Mn²⁺ as cofactors. These metal ions are essential for the enzyme’s hydrolytic activity on SAH, facilitating the regeneration of homocysteine and adenosine.
This overview highlights the critical role of SAM in methylation and the essential enzyme SAHH in regulating methylation balance by managing SAH levels.
Challenges to Naturalistic Explanations of S-Adenosylmethionine (SAM) Metabolism
1. Complex Enzymatic Reactions and Molecular Recognition: The synthesis and utilization of SAM involve highly specific enzymatic reactions that present significant challenges:
- How did enzymes like methionine adenosyltransferase (MAT) develop the ability to catalyze the formation of the high-energy sulfonium compound SAM?
- What intermediate forms, if any, could have existed that were functional in SAM synthesis?
- How did these enzymes acquire the precise molecular recognition capabilities required for substrate binding and catalysis?
2. Interdependence of Reactions in the SAM-Dependent Methylation Cycle: The cycle of SAM-dependent methylation presents a chicken-and-egg problem:
- How could the cycle have emerged when each step depends on the products of the previous reactions?
- What intermediate forms of this cycle, if any, could have been functional?
- How did the precise coordination between methyltransferases, SAH hydrolase, and methionine regeneration enzymes develop?
3. Cofactor Dependence and Methionine Regeneration: The regeneration of methionine involves complex enzymes and cofactors:
- How did the dependence on vitamin B12 and folate in methionine synthase develop?
- What were the intermediate steps, if any, in the emergence of betaine-homocysteine methyltransferase?
- How did these diverse cofactor requirements become integrated into a single metabolic pathway?
4. Multi-layered Regulation of SAM Metabolism: The tight regulation of SAM metabolism at multiple levels poses significant questions:
- How did such sophisticated regulatory mechanisms develop?
- What intermediate forms of regulation, if any, could have been functional?
- How did the various levels of control (allosteric, transcriptional, post-translational) become integrated?
5. Integration with Other Metabolic Pathways: The deep integration of SAM metabolism with numerous other cellular processes presents challenges:
- How did SAM metabolism become so intricately linked with the folate cycle, transsulfuration pathway, and polyamine synthesis?
- What intermediate stages, if any, could have existed in the development of these interconnections?
- How did the precise coordination required between these pathways arise?
6. Enzyme Specificity and Catalytic Efficiency: The enzymes involved in SAM metabolism display remarkable specificity and efficiency:
- How did these enzymes acquire their high degree of substrate specificity?
- What mechanisms allowed for the development of such catalytic efficiency?
- How do these enzymes maintain their function in the presence of structurally similar molecules?
7. Folate Metabolism and One-Carbon Transfer: The intricate folate metabolism pathway, crucial for SAM synthesis, presents its own set of challenges:
- How did the complex network of enzymes involved in folate metabolism arise?
- What intermediate forms, if any, could have existed in the development of one-carbon transfer reactions?
- How did the precise coordination between folate metabolism and SAM synthesis develop?
8. Compartmentalization and Transport of SAM Metabolites: The cellular compartmentalization of SAM metabolism components poses additional questions:
- How did the specific localization of SAM metabolism enzymes in different cellular compartments develop?
- What mechanisms allowed for the emergence of specific transporters for SAM and related metabolites?
- How did the precise coordination between compartmentalized reactions arise?
These challenges to naturalistic explanations of SAM metabolism highlight the need for further research and careful consideration of alternative hypotheses. The intricate nature of this system, its essential role in cellular function, and the complexity of its components raise significant questions about its origin and development.
10.12. Biotin Biosynthesis
Biotin biosynthesis represents a remarkable feat of biochemical engineering. This process involves a series of highly specific enzymatic reactions, each catalyzed by a unique enzyme with precise substrate recognition capabilities. The pathway begins with pimeloyl-CoA and progresses through several intermediates before culminating in the formation of biotin. The first step in this process involves the condensation of pimeloyl-CoA with L-alanine, catalyzed by 8-amino-7-oxononanoate synthase. This reaction requires precise molecular recognition and positioning of both substrates. The enzyme must overcome significant energetic barriers to form the carbon-nitrogen bond, a process that demands exquisite catalytic prowess. Subsequent steps in the pathway involve equally complex reactions. The conversion of 8-amino-7-oxononanoate to 7,8-diaminononanoate, catalyzed by 8-amino-7-oxononanoate aminotransferase, requires the transfer of an amino group from a donor molecule. This reaction demands not only substrate specificity but also the ability to facilitate the transfer of chemical groups between molecules. The formation of dethiobiotin from 7,8-diaminononanoate, catalyzed by dethiobiotin synthetase, involves the ATP-dependent closure of a ureido ring. This step represents a significant increase in molecular complexity, requiring precise control over the reaction trajectory to ensure the correct product is formed. The final step, the conversion of dethiobiotin to biotin by biotin synthase, is perhaps the most remarkable. This reaction involves the insertion of a sulfur atom into an unactivated carbon-hydrogen bond, a feat that pushes the boundaries of known biochemistry. The enzyme employs a complex iron-sulfur cluster and S-adenosyl methionine as a radical initiator to accomplish this challenging transformation.
10.12.1. Enzyme Specificity and Catalytic Efficiency
The enzymes involved in biotin biosynthesis exhibit remarkable substrate specificity and catalytic efficiency. Each enzyme in the pathway must recognize its specific substrate among a sea of structurally similar molecules within the cell. This level of discrimination requires precisely shaped binding pockets and intricate networks of chemical interactions between the enzyme and its substrate. Moreover, these enzymes catalyze their respective reactions with extraordinary efficiency. They accelerate reaction rates by factors of millions or even billions, allowing the cell to produce biotin on biologically relevant timescales. This catalytic prowess is achieved through complex mechanisms involving precisely positioned catalytic residues, controlled micro-environments within the active site, and dynamic conformational changes during the catalytic cycle.
10.12.2. Pathway Integration and Regulation
The biotin biosynthesis pathway does not exist in isolation but is intimately connected with other cellular processes. The pathway's starting material, pimeloyl-CoA, intersects with fatty acid metabolism. The pathway also connects with amino acid metabolism through the use of L-alanine and the aminotransferase reaction. Furthermore, the final step requires S-adenosyl methionine, linking biotin synthesis to one-carbon metabolism. This integration demands precise regulation to ensure that biotin production matches cellular needs without depleting resources required for other essential processes. The pathway is subject to complex regulatory mechanisms, including feedback inhibition and transcriptional control. These regulatory systems must have developed in concert with the biosynthetic pathway itself, adding another layer of complexity to the system.
10.12.3. Cofactor Dependence
Several steps in the biotin biosynthesis pathway require specific cofactors. The aminotransferase reaction depends on pyridoxal phosphate, while the final sulfur insertion step requires both an iron-sulfur cluster and S-adenosyl methionine. The dependence on these cofactors raises additional questions about the origin of the pathway. How did these enzymes develop their ability to bind and utilize these complex cofactors? How did the cell ensure the availability of these cofactors in concert with the development of the biotin synthesis pathway?
Biotin biosynthesis is a crucial metabolic pathway that produces biotin (vitamin B7), an essential cofactor for carboxylase enzymes involved in fatty acid synthesis, gluconeogenesis, and amino acid metabolism. This pathway is present in many bacteria, fungi, and plants, but most animals, including humans, lack the ability to synthesize biotin and must obtain it from their diet. The biotin biosynthesis pathway is of significant interest due to its potential as a target for antimicrobial drugs and its importance in industrial biotechnology.
Key enzymes involved:
Lysine 6-aminotransferase (EC 2.6.1.36): Smallest known: 405 amino acids (Thermus thermophilus). This enzyme catalyzes the first step in biotin biosynthesis, converting L-lysine to L-2,6-diaminopimelate. It plays a crucial role in initiating the pathway and is essential for organisms that synthesize biotin de novo.
7,8-Diaminononanoate synthase (EC 6.3.1.25): Smallest known: 384 amino acids (Aquifex aeolicus). This enzyme catalyzes the synthesis of 7,8-diaminononanoate from 7-keto-8-aminopelargonic acid and S-adenosyl methionine. It is critical for the formation of the carbon skeleton of biotin.
Dethiobiotin synthetase (EC 6.3.3.3): Smallest known: 224 amino acids (Helicobacter pylori). This enzyme catalyzes the formation of dethiobiotin from 7,8-diaminononanoate. It is essential for creating the ureido ring structure characteristic of biotin.
Biotin synthase (EC 2.8.1.6): Smallest known: 316 amino acids (Bacillus subtilis). This enzyme catalyzes the final step in biotin biosynthesis, converting dethiobiotin to biotin. It is crucial for completing the biotin structure and is often considered the rate-limiting step in the pathway.
The biotin biosynthesis essential enzyme group consists of 4 enzymes. The total number of amino acids for the smallest known versions of these enzymes is 1,329.
Information on metal clusters or cofactors:
Lysine 6-aminotransferase (EC 2.6.1.36): Requires pyridoxal 5'-phosphate (PLP) as a cofactor. PLP is covalently bound to a lysine residue in the active site and is essential for the transamination reaction.
7,8-Diaminononanoate synthase (EC 6.3.1.25): Requires ATP and Mg²⁺ for its catalytic activity. The magnesium ion acts as a cofactor, facilitating the ATP-dependent reaction.
Dethiobiotin synthetase (EC 6.3.3.3): Requires ATP and Mg²⁺ for its catalytic activity. The magnesium ion is essential for ATP binding and the subsequent reaction.
Biotin synthase (EC 2.8.1.6): Contains an iron-sulfur cluster ([4Fe-4S]) and requires S-adenosyl methionine (SAM) as a cofactor. The iron-sulfur cluster is crucial for the radical SAM mechanism used to insert the sulfur atom into dethiobiotin.
This overview highlights the complexity and importance of the biotin biosynthesis pathway, emphasizing the unique roles of each enzyme and their cofactor requirements. The pathway's absence in most animals makes it an attractive target for antimicrobial drug development, while its presence in certain microorganisms is leveraged for industrial biotin production.
Utilization of Biotin
Acetyl-CoA carboxylase: EC: 6.4.1.2 - Utilizes biotin to carboxylate acetyl-CoA to malonyl-CoA.
Recycling and Conversion of Biotin
Biotinidase: EC: 3.5.1.76 Hydrolyzes biocytin to release biotin for recycling.
Biotinidase: EC: 3.5.1.76 - Hydrolyzes biocytin to release biotin for recycling.
Challenges in Understanding Biotinidase Function and Regulation
Biotinidase exhibits remarkable specificity in recognizing and hydrolyzing biocytin. This presents several challenging questions:
- How did biotinidase develop its precise active site configuration to accommodate biocytin?
- What intermediate forms, if any, could have existed that were functional in biotin recycling?
- How does biotinidase distinguish biocytin from structurally similar molecules in the cellular milieu?
1. Catalytic Mechanism and Efficiency: The catalytic mechanism of biotinidase involves complex proton transfers and nucleophilic attack. This raises several questions:
- How did the precise arrangement of catalytic residues in biotinidase's active site arise?
- What is the exact sequence of chemical events during catalysis, and how is it coordinated?
- How does biotinidase achieve its high catalytic efficiency?
2. Regulation of Biotinidase Activity: The regulation of biotinidase activity is crucial for maintaining proper biotin levels. This presents several challenges:
- How is biotinidase activity coordinated with biotin synthesis and utilization?
- What mechanisms control biotinidase expression and activity in response to cellular biotin levels?
- How did these regulatory mechanisms develop in concert with biotinidase itself?
Studies by Pindolia et al. (2011) have shown that biotinidase expression is regulated by biotin status, but the molecular details of this regulation remain unclear.
4. Multifunctionality of Biotinidase: Biotinidase has been found to have functions beyond biotin recycling, including a potential role in processing biotinylated histones. This raises several questions:
- How did biotinidase acquire these additional functions?
- What is the relationship between biotinidase's different functions?
- How does the cell regulate these diverse activities?
5. Biotinidase Deficiency and Genetic Variations: Biotinidase deficiency is a genetic disorder with varying degrees of severity. This presents several challenges:
- How do different mutations in the biotinidase gene affect enzyme function?
- What is the relationship between enzyme structure and the various clinical presentations of biotinidase deficiency?
- How has biotinidase maintained its function despite genetic variations across populations?
The study of biotinidase presents numerous challenges that defy simple explanations. The enzyme's structural complexity, catalytic sophistication, regulatory mechanisms, and multifunctionality raise profound questions about its origins and development. Current research continues to uncover new aspects of biotinidase function, but many fundamental questions remain unanswered. Understanding these aspects fully will require interdisciplinary approaches and novel experimental techniques.
10.13. Carbon Monoxide Dehydrogenase (CODH): A Marvel of Biochemical Engineering
Carbon Monoxide Dehydrogenase (CODH) represents a remarkable feat of biochemical engineering, playing a crucial role in carbon cycling and autotrophic growth in certain microorganisms.
CODH is named for its primary function:
1. "Carbon Monoxide" refers to its substrate, CO.
2. "Dehydrogenase" indicates its role in removing hydrogen (in this case, as part of oxidizing CO to CO2).
The name reflects the enzyme's ability to catalyze the oxidation of carbon monoxide (CO) to carbon dioxide (CO2), effectively removing hydrogen from the substrate. While some CODHs can also catalyze the reverse reaction, the name emphasizes its historically first-discovered and most prominent function. This enzyme catalyzes the interconversion of carbon monoxide (CO) and carbon dioxide (CO2), a reaction central to the Wood-Ljungdahl carbon fixation pathway. The complexity and efficiency of CODH raises questions about its origin and function, challenging our understanding of biochemical systems. CODH exists in two main forms: the monofunctional CODH (EC: 1.2.99.2) and the bifunctional CODH/Acetyl-CoA Synthase (CODH/ACS) complex (EC: 1.2.7.4). The monofunctional CODH primarily oxidizes CO to CO2, while the bifunctional complex not only catalyzes this reaction but also participates in the synthesis of acetyl-CoA from CO2, CO, and a methyl group. These enzymes demonstrate extraordinary catalytic prowess, operating at the thermodynamic limit with minimal overpotential.
10.13.1. Catalytic Efficiency
CODHs are among the most efficient enzymes known, operating near the thermodynamic limit of the CO/CO2 interconversion reaction. This means they catalyze the reaction with minimal energy loss, achieving maximum possible efficiency. The turnover rates (kcat) of CODHs are impressively high:
- For CO oxidation: up to 40,000 s⁻¹
- For CO2 reduction: up to 12 s⁻¹
These rates are among the highest observed for metalloenzymes, especially considering the complexity of the reaction. CODHs operate with an overpotential of only about 90 mV for CO oxidation. This is remarkably low, especially when compared to synthetic catalysts which typically require overpotentials of 400-600 mV for similar reactions. Imagine you're trying to push a heavy box up a small hill. The hill represents the energy barrier that needs to be overcome for a chemical reaction to occur. In an ideal world, you'd only need to exert exactly enough energy to get the box to the top of the hill. This "ideal" amount of energy is like the theoretical minimum energy needed for a chemical reaction. Now, in reality, you might need to push a bit harder than this ideal minimum to get the box moving and overcome friction. This extra push is similar to what we call "overpotential" in chemistry. In the context of Carbon Monoxide Dehydrogenase (CODH) enzymes:
The "hill" is the energy barrier for converting CO to CO2.
The "ideal push" is the theoretical minimum energy (or voltage) needed to make this conversion happen.
The "extra push" (overpotential) is the additional energy the enzyme actually needs to use above this theoretical minimum.
When we say CODHs operate with an overpotential of only about 90 mV for CO oxidation, it means:
- These enzymes need only a tiny bit of extra energy (90 millivolts) beyond the theoretical minimum to catalyze the reaction.
- This is remarkably low - like needing only a small extra push to get that heavy box over the hill.
- Most artificial catalysts we've created for similar reactions need a much bigger "extra push" - often 4-6 times more (400-600 mV).
To put it in everyday terms, it's like CODH enzymes are incredibly efficient cars that can climb a hill using just a touch more gas than the absolute minimum required. In contrast, many of our artificial catalysts are like less efficient vehicles that need to rev their engines much harder to climb the same hill. This extremely low overpotential is one of the reasons why CODHs are considered so remarkably efficient. They're doing a complex chemical job with very little wasted energy.
10.13.2. Mechanisms of Efficiency
1. Optimized Active Site Structure: The [NiFe4S4] C-cluster is precisely arranged to facilitate electron transfer and substrate binding. The asymmetric position of the nickel ion allows for optimal interaction with CO and CO2. The [NiFe4S4] C-cluster at the heart of Carbon Monoxide Dehydrogenase (CODH) is indeed a marvel of biochemical engineering, showcasing an extraordinary level of complexity and precision. Let's break down the intricacies of this structure and its assembly:

a Crystal structure of ACS/CODH complex (Darnault et al. 2003). ACS forms the bifunctional enzyme with carbon monoxide dehydrogenase (CODH), which converts carbon dioxide into carbon monoxide, AcetylCoA Synthase/Carbon Monoxide Dehydrogenase (ACS/CODH). The structure of the CODH/ACS enzyme consists of the CODH enzyme as a dimer at the center with two ACS subunits on each side (Ragsdale 2004). b Structure of A-cluster (Svetlitchnyi et al. 2004). c Structure of C-cluster (Dobbek et al. 2001) 1
[/size]