41. Spatiotemporal Gene Expression
Spatiotemporal Gene Expression
Spatiotemporal gene expression refers to the precise regulation of gene activity in both space and time within an organism. It involves controlling which genes are turned on or off, and to what extent, in specific cells or tissues and at specific developmental stages or environmental conditions.
This level of gene expression regulation is of paramount importance in biological systems for several reasons:
Cell Specialization and Tissue Formation: During development, different cells within an organism acquire distinct identities and functions. This is achieved through precise spatiotemporal gene expression, allowing cells to differentiate into specific cell types and form tissues with specific functions.
Organismal Adaptation: Organisms need to adapt to changing environments and physiological demands. Spatiotemporal gene expression allows for the activation of genes that are relevant to the current conditions, ensuring survival and proper functioning.
Disease and Homeostasis: Dysregulation of spatiotemporal gene expression can lead to diseases. Proper regulation is crucial for maintaining homeostasis and preventing conditions like cancer, where genes are inappropriately expressed.
Developmental Patterning: The formation of complex body structures and organs requires precise gene expression patterns. Developmental genes are activated or repressed in specific regions and at specific times to shape the overall form of the organism.
Developmental Processes Shaping Organismal Form and Function
Developmental processes are a series of orchestrated events that shape the form and function of organisms as they grow and mature. These processes encompass a wide range of events, from cell differentiation and tissue formation to the establishment of body axes and the growth of complex structures. The interactions between various intracellular and extracellular systems are essential for these processes:
Cell Differentiation: As an organism develops, stem cells differentiate into specialized cell types with specific functions. Intracellular signaling and gene expression play a critical role in guiding cells towards their designated roles.
Morphogenesis: Morphogenesis is the process by which tissues and organs take on their distinct shapes. Intracellular mechanisms like cytoskeleton dynamics and extracellular cues from the environment guide this process.
Pattern Formation: The establishment of body axes and spatial patterns is crucial for proper development. Signaling gradients, gene expression gradients, and intercellular communication contribute to this patterning.
Growth and Organogenesis: Coordinated growth of tissues and organs ensures that an organism develops to its proper size and proportions. Growth factors, hormonal signaling, and nutrient availability are involved in this process.
Cell-Cell Communication and Signaling: Extracellular signaling molecules, including growth factors and morphogens, guide cellular behaviors and coordinate developmental processes between different cell types.
Genetic Regulation: Spatiotemporal gene expression controls the activation of specific genes during different developmental stages and in different parts of the organism, ensuring proper development and function.
Overall, developmental processes involve intricate coordination between various systems, both within cells and in their surrounding environment. This orchestration ensures that organisms develop correctly, with the appropriate structures, functions, and adaptations to their environment.
Orchestrating precise gene expression in time and space
The orchestration of precise gene expression in both time and space is a fundamental aspect of biological systems. This dynamic regulation ensures that genes are activated or repressed with exquisite precision, enabling cells and organisms to respond appropriately to their developmental stage, environmental cues, and physiological needs.
Temporal Gene Expression: Temporal regulation of gene expression involves controlling when specific genes are activated or silenced during an organism's lifecycle. This temporal orchestration is vital for processes such as embryonic development, tissue repair, and circadian rhythms. Genes must be turned on or off at the right moments to ensure the proper progression of events.
Spatial Gene Expression: Spatial regulation determines where genes are expressed within an organism. Different cells and tissues have distinct roles, and precise gene expression localization is crucial for achieving cellular specialization and forming complex structures. This spatial control ensures that cells in different regions adopt specific identities and functions.
Signaling Pathways: Cellular signaling pathways play a crucial role in spatiotemporal gene expression. Signaling molecules transmit information from the extracellular environment to the cell's nucleus, where they influence gene transcription. This allows cells to interpret their surroundings and adjust gene expression accordingly.
Transcription Factors: Transcription factors are proteins that bind to specific DNA sequences and regulate gene expression. Their presence or absence can turn genes on or off, and their activity can be influenced by various factors, including signaling pathways and cellular context.
Epigenetic Modifications: Epigenetic modifications, such as DNA methylation and histone modifications, play a role in regulating gene expression patterns. These modifications can influence the accessibility of genes to transcription machinery, thereby impacting their activity.
Developmental Processes: During development, genes are expressed in specific patterns that guide the formation of tissues, organs, and body structures. This requires precise spatiotemporal coordination to ensure that genes are activated or repressed in the right places and at the right times.
Environmental Responses: Cells also adjust their gene expression patterns in response to environmental changes. This adaptability allows organisms to survive and thrive in different conditions by activating genes that provide a selective advantage.
Disease and Dysfunction: Dysregulation of spatiotemporal gene expression can lead to diseases, including developmental disorders and cancer. Understanding the underlying mechanisms of precise gene regulation is essential for deciphering disease origins and developing targeted therapies.
Orchestrating precise gene expression in time and space is a remarkable feat of biological complexity. This regulation enables organisms to navigate intricate developmental pathways, respond to changing environments, and maintain proper function. The interplay of signaling, transcription factors, epigenetic modifications, and other molecular processes ensures that genes are turned on or off with remarkable precision, ultimately shaping the remarkable diversity and functionality of life.
Mechanisms governing spatiotemporal gene patterns in diverse processes
The establishment and maintenance of precise spatiotemporal gene expression patterns are governed by a complex interplay of mechanisms that contribute to the diversity of biological processes. These mechanisms ensure that genes are activated or repressed at specific times and in specific locations, playing critical roles in development, adaptation, and cellular function.
Transcriptional Regulation: Transcription factors and enhancers are key players in spatiotemporal gene expression. Transcription factors bind to DNA sequences and activate or suppress gene transcription. Enhancers are DNA elements that can be far from the gene they regulate and interact with transcription factors to fine-tune gene expression in specific contexts.
Epigenetic Modifications: Epigenetic marks, such as DNA methylation and histone modifications, influence gene accessibility. These modifications can act as "marks" that determine whether a gene is actively transcribed or silenced, contributing to spatiotemporal regulation.
Chromatin Remodeling: Chromatin structure can be altered by chromatin remodeling complexes, making genes more or less accessible to transcription machinery. This dynamic alteration plays a role in controlling gene expression patterns.
Non-Coding RNAs: Non-coding RNAs, including microRNAs and long non-coding RNAs, can regulate gene expression by binding to target messenger RNAs (mRNAs) and affecting their stability or translation. They contribute to fine-tuning gene expression in specific tissues or developmental stages.
Cellular Signaling Pathways: Extracellular signals, transmitted through signaling pathways, influence gene expression. Activation of specific pathways in response to environmental cues or developmental signals can lead to changes in gene expression patterns.
Feedback Loops: Feedback loops involve regulatory proteins that control their own expression. These loops contribute to maintaining stable gene expression patterns and responding to fluctuations in cellular conditions.
Splicing and Alternative Promoters: Different exons can be included or excluded from mRNAs through alternative splicing, resulting in protein isoforms with distinct functions. Alternative promoters can also drive tissue-specific gene expression by producing different mRNA variants.
Nuclear Organization: The three-dimensional organization of the nucleus can influence gene expression. Genes located in close proximity within the nucleus may share regulatory elements, affecting their coordinated expression.
Cell-Cell Communication: Cells communicate with each other through signaling molecules that influence neighboring cells' gene expression patterns. This communication is crucial for coordinating development and maintaining tissue integrity.
Feedback and Feedforward Networks: Regulatory networks involving multiple genes can create feedback and feedforward loops, which contribute to robust and coordinated gene expression patterns.
The intricate interplay of these mechanisms allows organisms to generate a wide range of spatiotemporal gene expression patterns, facilitating processes such as embryonic development, tissue regeneration, immune responses, and adaptation to changing environments. The diversity of these mechanisms underscores the complexity of life's regulatory processes and highlights the remarkable precision with which genes are controlled to ensure optimal function.
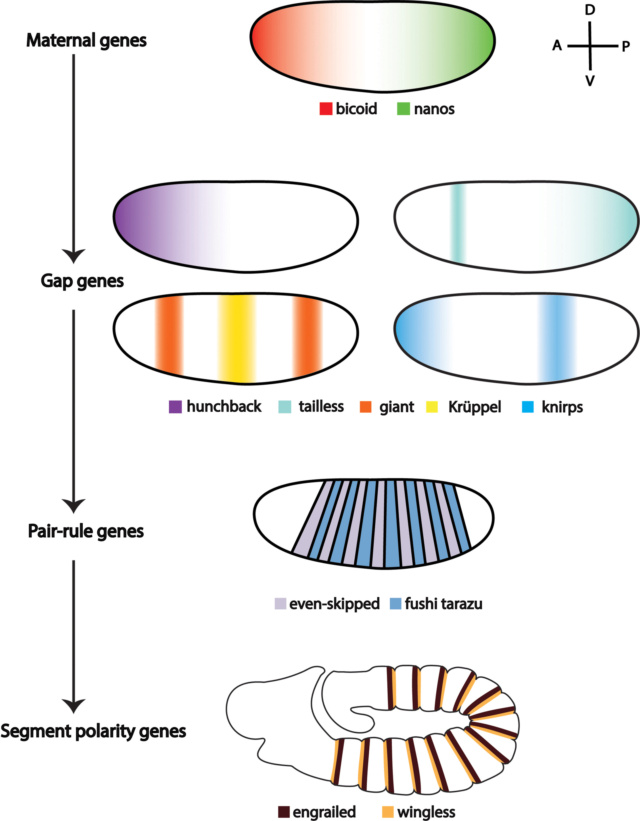
AP patterning starts with maternal coordinate genes that regulate the expression of downstream zygotic genes in a hierarchic fashion. The maternal factors Bicoid and Nanos activate the expression of zygotic gap genes (hunchback, tailless, giant, Krüppel, knirps, and others). The products of the gap genes, combined with maternal morphogens, in turn, control the expression of pair-rule genes in seven stripes (e.g., even-skipped and fushi tarazu). Finally, the AP polarity of the parasegments is controlled by segment polarity genes, among which wingless (wg) and engrailed (en) are expressed along the segmental boundaries. 1
Deciphering the evolutionary timeline of spatiotemporal gene controls
The evolution of spatiotemporal gene controls would have unfolded over millions of years, driven by the gradual emergence of complex regulatory mechanisms that enabled organisms to adapt to diverse environments and developmental demands.
Early Gene Expression Patterns: In the earliest stages of life, simple organisms would have exhibited basic gene expression patterns that responded to fundamental environmental cues. These patterns would have been essential for survival and reproduction but lacked the intricate spatiotemporal specificity seen in more complex organisms.
Multicellular Evolution and Tissue Formation: With the emergence of multicellularity, the need for specialized cell types and tissues would have driven the development of more refined gene expression controls. Basic spatial patterns of gene expression would have been established to coordinate cell differentiation and contribute to the functionality of distinct cell populations.
Spatial Patterning in Development: As organisms diversified and developed more complex body plans, the establishment of precise spatial gene expression patterns would have become crucial. Regulatory mechanisms would have evolved to ensure that genes are activated or repressed in specific regions, contributing to the formation of intricate structures and organs.
Evolution of Signaling Pathways: The emergence of sophisticated signaling pathways and networks would have allowed organisms to respond to a wider range of environmental cues. These pathways would have interacted with gene regulatory elements, enabling cells to fine-tune their responses based on spatial and temporal inputs.
Cellular Differentiation and Specialization: As organisms evolved, more specialized cell types and tissues would have developed. This required the evolution of mechanisms that finely control gene expression to maintain cell identity and function within specific spatial contexts.
Fine-Tuning Developmental Timing: The evolution of spatiotemporal gene controls would also encompass the regulation of developmental timing. Genes would have been activated or repressed in specific sequences and at specific stages to ensure the proper progression of developmental events.
Adaptation and Environmental Responses: Organisms would have faced changing environments and selective pressures. The evolution of spatiotemporal gene controls would have allowed for the adaptation of gene expression patterns in response to new ecological niches, optimizing survival and reproduction.
Increased Complexity and Diversity: As evolution continued, organisms with more intricate and versatile spatiotemporal gene control mechanisms would have gained a competitive edge. This would have driven the diversification of developmental strategies and the emergence of complex life forms.
Co-Evolution of Mechanisms: The various mechanisms involved in spatiotemporal gene control, such as transcriptional regulation, epigenetic modifications, and cellular signaling, would have co-evolved to ensure their compatibility and effectiveness in coordinating gene expression.
The evolution of spatiotemporal gene controls represents a gradual process intertwined with the increasing complexity and diversity of life forms. These mechanisms would have played a pivotal role in shaping the development, adaptation, and functionality of organisms, contributing to the remarkable biological diversity observed in the natural world.
Genetic Components Essential for Spatiotemporal Gene Regulation
Spatiotemporal gene regulation is a complex process that involves the interplay of various genetic components, each contributing to the precise control of gene expression patterns in different regions and at different times within an organism. These components ensure the proper development, function, and adaptation of cells and tissues.
Promoters and Enhancers: Promoters are DNA sequences located near the start of a gene that serve as binding sites for RNA polymerase, initiating transcription. Enhancers are regulatory DNA elements that can be located far from the gene they regulate. They interact with transcription factors and other regulatory proteins to enhance or suppress gene expression in specific contexts.
Transcription Factors: Transcription factors are proteins that bind to DNA sequences and influence gene expression. They include activators that promote transcription and repressors that inhibit it. The presence and activity of specific transcription factors determine which genes are expressed and when.
Cis-Regulatory Modules: Cis-regulatory modules are combinations of enhancers and promoters that work together to regulate gene expression. These modules can contain multiple binding sites for different transcription factors, allowing for precise spatiotemporal control.
Epigenetic Marks: Epigenetic modifications, such as DNA methylation and histone modifications, influence gene accessibility. These marks can be inherited through cell divisions and contribute to the establishment of stable gene expression patterns.
Chromatin Remodeling Complexes: These complexes alter the structure of chromatin, making genes more or less accessible to transcription machinery. They play a role in fine-tuning gene expression patterns by controlling the availability of regulatory elements.
Non-Coding RNAs: Non-coding RNAs, including microRNAs and long non-coding RNAs, regulate gene expression by binding to target mRNAs. They can influence mRNA stability or translation efficiency, contributing to spatiotemporal regulation.
Cellular Signaling Pathways: Signaling pathways transmit information from the extracellular environment to the nucleus. They can activate or inhibit transcription factors, influencing gene expression based on developmental cues or environmental conditions.
Nuclear Organization: The three-dimensional organization of the nucleus can affect gene expression. Genes located in close proximity within the nucleus may share regulatory elements, allowing for coordinated expression.
Alternative Splicing: Many genes undergo alternative splicing, producing multiple mRNA isoforms from a single gene. This diversifies protein products and contributes to tissue-specific gene expression.
Feedback and Feedforward Loops: Regulatory networks involving multiple genes can create feedback and feedforward loops that contribute to robust and coordinated gene expression patterns.
Evolutionarily Conserved Elements: Some genetic elements are conserved across species and play critical roles in spatiotemporal gene regulation. These elements are often indicative of their importance in fundamental developmental processes.
Genome Accessibility Factors: Factors that influence chromatin accessibility, such as chromatin remodelers and pioneer factors, ensure that genes can be transcribed when needed.
Chromatin Boundary Elements: These elements define the boundaries of regulatory domains, preventing the spread of regulatory signals to inappropriate genes.
Adaptive Evolution of Regulatory Elements: As organisms adapt to new environments, the genetic components involved in spatiotemporal gene regulation can evolve to optimize gene expression for the organism's survival and fitness.
The coordination and interaction of these genetic components create a sophisticated regulatory network that enables precise spatiotemporal control of gene expression, allowing organisms to develop, function, and adapt in complex and dynamic environments.
Manufacturing codes steering spatiotemporal gene expression
The intricate process of spatiotemporal gene expression can be likened to the execution of manufacturing codes that guide the precise activation or repression of genes in specific spatial and temporal contexts. These "codes" involve a complex interplay of molecular components that work together to orchestrate the developmental and functional processes within an organism.
Genetic Blueprint: The DNA sequence itself serves as the foundational blueprint containing the instructions for building and regulating an organism. Specific sequences act as promoters, enhancers, and regulatory elements that are essential for controlling gene expression.
Transcription Factors as Programmers: Transcription factors can be thought of as programmers that read the genetic code and interpret it to initiate or suppress gene transcription. These proteins recognize specific DNA sequences and bind to them, initiating the transcriptional machinery.
Enhancers as Activation Codes: Enhancers can be likened to activation codes that, when bound by transcription factors, enhance gene expression. The presence of particular combinations of transcription factors at enhancers determines the level and timing of gene activation.
Repressors as Inhibitory Codes: Repressors function as inhibitory codes that prevent gene expression. They can bind to DNA sequences or interfere with the activity of transcription factors, ensuring that certain genes remain inactive in specific cellular contexts.
Epigenetic "Locks" and "Keys": Epigenetic modifications can be likened to locks and keys that regulate access to genes. Methylation and histone modifications serve as locks, rendering genes inaccessible. Conversely, specific transcription factors and co-activators act as keys that unlock genes for transcription.
Chromatin Remodeling as Assembly Lines: Chromatin remodeling complexes can be envisioned as assembly lines that modify the structure of chromatin, making genes more accessible or repressed. These complexes modify the chromatin landscape, allowing for the execution of gene expression codes.
Cell Signaling as Communication Protocols: Cellular signaling pathways function as communication protocols that relay external cues to the nucleus. They convey information about environmental conditions and developmental stages, guiding the execution of appropriate gene expression programs.
Non-Coding RNAs as Messengers: Non-coding RNAs act as messengers that transmit regulatory information to target genes. MicroRNAs and long non-coding RNAs can interact with mRNA molecules, influencing their stability and translation efficiency.
Evolution as Iterative Design Process: The evolutionary process can be seen as an iterative design process that refines the manufacturing codes over generations. Beneficial changes in regulatory elements and transcription factor binding sites are selected for, optimizing the precision and adaptability of gene expression.
Cellular Identity and Specialization as Output: The culmination of these manufacturing codes is the establishment of cellular identity and specialization. Cells "read" the codes to determine their roles and functions within the organism, contributing to the overall structure and functionality of tissues and organs.
Just as manufacturing codes ensure the proper assembly and functioning of complex machinery, the intricate genetic components, and mechanisms that govern spatiotemporal gene expression ensure the precise development, adaptation, and coordination of biological systems in the exquisite tapestry of life.
Epigenetic Tools Modulating Timely Gene Activations
Epigenetic mechanisms play a crucial role in regulating the timing of gene activations, contributing to the precise orchestration of gene expression patterns during development and in response to various environmental cues. These epigenetic tools provide a dynamic and adaptable framework for ensuring that genes are activated at the right time and in the right context.
DNA Methylation: DNA methylation involves the addition of methyl groups to cytosine bases, often leading to gene repression. During development, specific genes may be temporarily methylated, preventing their expression at inappropriate stages. Conversely, demethylation events can "unleash" genes when the time is right for their activation.
Histone Modifications: Histone modifications, such as acetylation, methylation, and phosphorylation, influence chromatin structure and gene accessibility. These modifications serve as epigenetic marks that can promote or inhibit transcription. Enzymes that add or remove these marks act as tools for regulating gene activation timing.
Chromatin Remodeling Complexes: Chromatin remodeling complexes utilize energy to reposition, evict, or alter nucleosomes, affecting the accessibility of genes to transcription machinery. These complexes act as tools for modifying chromatin structure and allowing genes to be activated or repressed when needed.
Long Non-Coding RNAs (lncRNAs): Long non-coding RNAs can interact with chromatin-modifying complexes and serve as guides for their recruitment to specific genomic regions. They act as epigenetic tools that contribute to the establishment of proper gene expression patterns during development.
Polycomb Group Proteins: Polycomb group proteins form complexes that regulate the maintenance of gene silencing during development. These proteins serve as epigenetic tools that ensure certain genes remain inactive until the appropriate developmental stage, preventing premature activation.
Chromatin States and Bivalent Domains: Certain genomic regions exhibit "bivalent" domains that possess both activating and repressive histone marks. These domains keep critical developmental genes poised for activation, allowing for rapid responses to cues while maintaining repression until needed.
Developmental Clocks and Timers: Epigenetic changes can be linked to internal biological clocks that regulate gene expression timing. Circadian rhythms, for instance, involve epigenetic modifications that enable genes to be activated or repressed according to specific time windows.
Environmental Sensing and Adaptation: Epigenetic modifications can be influenced by environmental factors, allowing organisms to adapt their gene expression patterns in response to changing conditions. This epigenetic plasticity acts as a tool for timely adjustments to various stimuli.
Evolutionary Conservation of Epigenetic Mechanisms: The conservation of epigenetic tools across species suggests their importance in regulating timely gene activations. These mechanisms have been refined over evolutionary time to optimize the timing of gene expression for the development and survival of organisms.
Overall, epigenetic tools serve as intricate mechanisms for modulating timely gene activations. They allow organisms to fine-tune their gene expression patterns in response to developmental cues, environmental changes, and physiological demands, ensuring that genes are activated or repressed at the appropriate moments for optimal function.
Interplay between Signaling Pathways and Gene Expression Patterns
The interplay between signaling pathways and gene expression patterns is a dynamic and intricate process that governs how cells and organisms respond to their environment, regulate development, and maintain homeostasis. This coordination ensures that cellular activities are precisely controlled and adapted to changing conditions.
Sensory Perception and Signal Initiation: Signaling pathways are initiated by sensory inputs, such as growth factors, hormones, or environmental cues. These signals are detected by receptors on the cell surface or within the cell, triggering a cascade of events.
Transduction and Signal Amplification: Once a signal is detected, it is transduced into a series of biochemical events within the cell. This often involves amplification of the signal, where a single extracellular ligand can lead to the activation of multiple intracellular molecules.
Activation of Transcription Factors: Signaling pathways frequently converge on transcription factors. Activated transcription factors are then transported to the nucleus, where they bind to specific DNA sequences, promoting or repressing gene transcription.
Gene Expression Changes: The binding of transcription factors to DNA directly influences gene expression. Genes targeted by transcription factors associated with a specific signaling pathway will be activated or repressed based on the pathway's activation status.
Fine-Tuning Gene Regulation: Multiple signaling pathways can converge on a single gene, providing layers of regulation. This fine-tuning ensures that gene expression is responsive to a combination of signals, allowing for more precise control.
Feedback Loops: Signaling pathways can regulate their own activity through feedback loops. Genes activated by a signaling pathway can encode proteins that modulate the pathway itself, leading to self-regulation.
Cross-Talk between Pathways: Signaling pathways can cross-talk, where components of one pathway influence the activity of another. This integration allows cells to integrate multiple inputs and generate a coordinated response.
Developmental Patterning: Signaling pathways are instrumental in establishing developmental patterns. During embryogenesis, for instance, gradients of signaling molecules can lead to distinct gene expression patterns that define tissue identities.
Environmental Adaptation: Signaling pathways enable cells to respond to changes in their environment. Cells can adjust their gene expression patterns to cope with varying conditions, optimizing their survival and function.
Dysregulation and Disease: Dysregulation of signaling pathways can lead to diseases. Aberrant activation of oncogenic signaling pathways can result in uncontrolled cell growth and cancer. Similarly, improper signaling can contribute to developmental disorders.
Evolutionary Adaptation: Signaling pathways have evolved to allow organisms to adapt to different environments and challenges. Evolutionary changes in signaling components can lead to modified gene expression patterns that confer selective advantages.
The intricate interplay between signaling pathways and gene expression patterns forms a complex regulatory network that ensures cells respond appropriately to their surroundings, guiding their development, function, and adaptation. This interwoven system highlights the remarkable coordination required for cells and organisms to thrive in diverse and ever-changing environments.
Regulatory Systems Harmonizing Spatiotemporal Gene Expression
The harmonization of spatiotemporal gene expression is orchestrated by a sophisticated network of regulatory systems that work in concert to ensure precise and coordinated gene activation or repression in specific spatial and temporal contexts. This orchestration is fundamental for proper development, functionality, and adaptability within complex biological systems.
Transcriptional Regulation and Enhancer-Promoter Interactions: Transcription factors and enhancers play a pivotal role in coordinating spatiotemporal gene expression. Transcription factors bind to enhancers and promoters, facilitating communication between distal regulatory elements and gene promoters, ensuring genes are activated or repressed when and where needed.
Epigenetic Mechanisms and Chromatin Remodeling: Epigenetic modifications and chromatin remodeling contribute to the establishment of permissive or repressive chromatin states. These mechanisms enable genes to be accessible or inaccessible to the transcription machinery, fine-tuning gene expression patterns across different cellular contexts.
Cellular Signaling Pathways and Signal Integration: Cellular signaling pathways converge on transcription factors, influencing their activity based on external cues. Integration of multiple signaling inputs ensures that genes are activated or repressed in response to a combination of signals, leading to a coordinated cellular response.
Temporal Regulation and Developmental Clocks: Biological clocks and developmental timers dictate the timing of gene expression. These internal timing mechanisms synchronize with external cues to activate or repress genes at specific developmental stages or circadian rhythms.
Spatial Patterning and Morphogen Gradients: Morphogen gradients establish spatial patterns of gene expression during embryogenesis. The concentration of signaling molecules provides positional information that guides the activation or repression of genes, contributing to the formation of distinct tissue identities.
Feedback and Feedforward Loops: Regulatory networks often incorporate feedback and feedforward loops. These loops maintain stable gene expression patterns and enable rapid responses to changes in cellular conditions, ensuring robustness and precision.
Cell-Cell Communication and Signaling Crosstalk: Cells communicate and coordinate their gene expression through signaling molecules. Signaling crosstalk allows neighboring cells to influence each other's gene expression patterns, contributing to the organization and function of tissues.
Alternative Splicing and Isoform Diversity: Alternative splicing generates different mRNA isoforms from a single gene, resulting in protein diversity. This mechanism contributes to the specialization of cell types and tissues through the production of distinct protein isoforms.
Evolutionary Conservation and Adaptation: Regulatory systems controlling spatiotemporal gene expression are evolutionarily conserved across species. While core mechanisms remain similar, variations allow organisms to adapt and optimize gene expression for their specific needs and environments.
Multilevel Integration and Robustness: The integration of various regulatory systems ensures the robustness of spatiotemporal gene expression. This multilevel coordination minimizes errors and deviations, maintaining the fidelity of developmental and physiological processes.
The intricate interplay of these regulatory systems creates a symphony of gene expression that harmonizes across different spatial and temporal scales, culminating in the intricate diversity and functionality observed within living organisms. This precise orchestration is essential for shaping the complexity of life and enabling organisms to thrive in diverse ecological niches.
Is there evidence for evolutionary mechanisms instantiating intricate gene expression controls?
Gene expression is an immensely intricate process. Every step in this process, from DNA transcription to protein synthesis, requires specific codes, signaling pathways, and molecular machinery. For the machinery to function properly, each of its many components must be precisely coordinated, leading many to question how such complexity could arise step-by-step through evolution. Consider the process of transcription, wherein a specific segment of DNA is read by RNA polymerase and transcribed into a complementary mRNA strand. This requires specific promoter regions on the DNA to signal the start of transcription and various transcription factors that recognize and bind to these promoters. The mere presence of these promoter regions and the specific binding proteins suggests an inherent coordination. Without these transcription factors, the promoters are useless. Conversely, without the promoters, the transcription factors have nothing to bind to. It's difficult to envision how one could evolve without the other, as each would bear no function on its own. Furthermore, the genetic code itself is a marvel of complexity. Each three-nucleotide codon on mRNA specifies a particular amino acid, a language that is then read and translated by ribosomes. This decoding process requires the precise interaction of mRNA with tRNA molecules that carry the appropriate amino acid. For this to function, not only does the mRNA codon need to exist and be transcribed correctly, but there also needs to be a corresponding tRNA molecule with the correct anticodon and attached amino acid, as well as the enzyme to attach that amino acid to the tRNA. The simultaneous existence and functional integration of all these parts seem improbable to have evolved in isolation. Then, there's the post-translational modification of proteins. Many proteins, once synthesized, undergo modifications like phosphorylation, methylation, or glycosylation. These modifications are essential for the protein's function, stability, or localization. The enzymes that carry out these modifications must recognize specific motifs on the proteins. Again, this implies a coordinated existence. The motif on the protein is pointless without the modifying enzyme, and the enzyme has no function without the protein motif.
Lastly, cellular signaling pathways show a level of interdependence and specificity that's breathtaking. For a signal to be transduced from the cell membrane to the nucleus, or between cells, numerous proteins and molecules need to interact in a highly regulated sequence. Missing just one component or having one non-functional component can disrupt the entire pathway. Given the intricacies of gene expression, it's hard to imagine how the multitude of processes and their respective molecular players could have evolved piecemeal. They seem to necessitate a complete, fully operational system from the onset to function efficiently and with purpose. Without this full system in place, it seems unlikely that partial or transitional stages would confer any evolutionary advantage, leading many to propose that such complexity and interdependence may suggest the work of an intelligent architect.
Investigating spatiotemporal gene controls for irreducibility traits
The complexity and precision observed in the realm of spatiotemporal gene controls present fascinating insights into the depth of coordination required for cellular operations. These processes can be described as complex mechanisms that could be viewed as both irreducible and interdependent.
Transcriptional Machinery: This includes the DNA itself, RNA polymerase, transcription factors, and promoter regions on DNA. Without each of these components interacting in harmony, the transcription of DNA into mRNA would be impossible.
Genetic Code: The codons of mRNA, the anticodons of tRNA, and the amino acids they attach to all have to match perfectly. A mistake here would result in the wrong protein being synthesized.
Ribosomal Translation: Ribosomes, along with mRNA and tRNA, translate the genetic code into proteins. Without the precise interaction between these molecules, protein synthesis is halted.
Post-translational Modifications: Many proteins require modifications after being synthesized. These modifications often require specific enzymes that recognize certain motifs on the proteins. The motifs and the enzymes are useless without each other.
Cellular Signaling: From ligands binding to receptors on the cell membrane, to secondary messengers, to protein kinases, every component in a signaling pathway is essential. Without one component, the signal cannot be relayed.
DNA Replication and Repair: DNA polymerases, helicases, topoisomerases, and various repair enzymes ensure DNA is replicated and any errors are fixed. A malfunction in one could be catastrophic for the cell.
Epigenetic Regulation: Histone modifications, DNA methylation, and non-coding RNAs all play roles in determining when and how genes are expressed. The language they 'speak' ensures that genes are expressed at the right time and in the right cells.
All these systems exhibit a form of irreducibility and interdependence. For example, in the case of the transcriptional machinery, transcription factors need promoters to bind to, while the promoters require the transcription factors for recognition. In terms of cellular signaling, a ligand without a receptor, or a kinase without a substrate, is functionally redundant. The codes and languages governing these processes crosstalk and integrate. DNA methylation might guide transcription factors, and ligands might trigger a cascade affecting gene expression. Given the level of coordination and the interplay between these systems, it's challenging to understand how such processes could evolve in isolation. The half-formed or transitional stages of such systems might not confer any evolutionary advantage, suggesting that such intricate systems, codes, and languages would need to be in place from the outset for efficient and purposeful function. The existence of such coordinated complexity may lead some to believe that they are the outcome of deliberate design.
Coordinated interactions post-establishment of spatiotemporal gene systems
The establishment of spatiotemporal gene systems ensures that genes are expressed in the right cells, at the right time, and in response to the right stimuli. But the complexity doesn't end there. Post-establishment of these gene systems, an intricate array of coordinated interactions is essential to maintain, modulate, and respond to the cellular environment. These interactions, functioning as a symphony of molecular events, are foundational to the robust and dynamic nature of cellular life.
Signal Transduction: Cells are in constant dialogue with their surroundings. External signals, whether they be hormones, nutrients, or other molecules, bind to cell receptors, triggering a cascade of intracellular events. These pathways often converge on transcriptional regulators, modifying gene expression in response to external cues.
Feedback Loops
Negative Feedback: Often, the product of a gene's expression will inhibit further expression of that gene. This ensures that cells don't overproduce certain proteins and maintain homeostasis.
Positive Feedback: Conversely, in some cases, the product of a gene might enhance its further expression. This can be crucial for rapid responses, such as in immune activation.
Tissue-specific Gene Regulation
Cell-to-Cell Communication: Cells within tissues don't act in isolation. Through mechanisms like paracrine signaling, one cell can influence the gene expression of its neighbors.
Epigenetic Landscaping: Epigenetic markers, influenced by both internal cellular states and external factors, play a significant role in tissue-specific gene expression. As cells differentiate, certain genes are 'locked' into on or off states, ensuring that cells maintain their specific identities.
Integration with Metabolic Pathways
Metabolite Regulation: Metabolic byproducts can directly influence gene expression. For example, when energy is abundant, high levels of ATP can impact the genes related to energy storage.
Temporal Regulation and Circadian Rhythms
Circadian Clocks: Many organisms have evolved internal clocks that regulate gene expression in 24-hour cycles. This ensures that certain processes, like DNA repair or photosynthesis, occur at optimal times of the day.
These multifaceted interactions underscore a profound interconnectedness within cellular systems. The regulation and coordination don't merely rest on establishing the spatiotemporal gene systems but persist throughout the cell's lifecycle. The exquisite precision and synchronized dance of these molecular interactions showcase the depth of complexity within even the simplest of cells. This might lead some to marvel at the ingenuity of such a system, wondering if such intricate coordination is the product of a specific design.
1. All systems that rely on semiotic code, languages, are interdependent and must emerge together interlocked, exhibit characteristics of deliberate design.
2. The processes of gene expression, transcription, translation, and post-translational modifications rely on semiotic code, languages, are interdependent, and must emerge together interlocked.
3. Therefore, the processes of gene expression, transcription, translation, and post-translational modifications exhibit characteristics of deliberate design.
1. Gheisari, E., Aakhte, M., & Müller, H.-A. J. (2020). Gastrulation in Drosophila melanogaster: Genetic control, cellular basis and biomechanics. Mechanisms of Development, 163, 103629. https://doi.org/10.1016/j.mod.2020.103629
Spatiotemporal Gene Expression
Spatiotemporal gene expression refers to the precise regulation of gene activity in both space and time within an organism. It involves controlling which genes are turned on or off, and to what extent, in specific cells or tissues and at specific developmental stages or environmental conditions.
This level of gene expression regulation is of paramount importance in biological systems for several reasons:
Cell Specialization and Tissue Formation: During development, different cells within an organism acquire distinct identities and functions. This is achieved through precise spatiotemporal gene expression, allowing cells to differentiate into specific cell types and form tissues with specific functions.
Organismal Adaptation: Organisms need to adapt to changing environments and physiological demands. Spatiotemporal gene expression allows for the activation of genes that are relevant to the current conditions, ensuring survival and proper functioning.
Disease and Homeostasis: Dysregulation of spatiotemporal gene expression can lead to diseases. Proper regulation is crucial for maintaining homeostasis and preventing conditions like cancer, where genes are inappropriately expressed.
Developmental Patterning: The formation of complex body structures and organs requires precise gene expression patterns. Developmental genes are activated or repressed in specific regions and at specific times to shape the overall form of the organism.
Developmental Processes Shaping Organismal Form and Function
Developmental processes are a series of orchestrated events that shape the form and function of organisms as they grow and mature. These processes encompass a wide range of events, from cell differentiation and tissue formation to the establishment of body axes and the growth of complex structures. The interactions between various intracellular and extracellular systems are essential for these processes:
Cell Differentiation: As an organism develops, stem cells differentiate into specialized cell types with specific functions. Intracellular signaling and gene expression play a critical role in guiding cells towards their designated roles.
Morphogenesis: Morphogenesis is the process by which tissues and organs take on their distinct shapes. Intracellular mechanisms like cytoskeleton dynamics and extracellular cues from the environment guide this process.
Pattern Formation: The establishment of body axes and spatial patterns is crucial for proper development. Signaling gradients, gene expression gradients, and intercellular communication contribute to this patterning.
Growth and Organogenesis: Coordinated growth of tissues and organs ensures that an organism develops to its proper size and proportions. Growth factors, hormonal signaling, and nutrient availability are involved in this process.
Cell-Cell Communication and Signaling: Extracellular signaling molecules, including growth factors and morphogens, guide cellular behaviors and coordinate developmental processes between different cell types.
Genetic Regulation: Spatiotemporal gene expression controls the activation of specific genes during different developmental stages and in different parts of the organism, ensuring proper development and function.
Overall, developmental processes involve intricate coordination between various systems, both within cells and in their surrounding environment. This orchestration ensures that organisms develop correctly, with the appropriate structures, functions, and adaptations to their environment.
Orchestrating precise gene expression in time and space
The orchestration of precise gene expression in both time and space is a fundamental aspect of biological systems. This dynamic regulation ensures that genes are activated or repressed with exquisite precision, enabling cells and organisms to respond appropriately to their developmental stage, environmental cues, and physiological needs.
Temporal Gene Expression: Temporal regulation of gene expression involves controlling when specific genes are activated or silenced during an organism's lifecycle. This temporal orchestration is vital for processes such as embryonic development, tissue repair, and circadian rhythms. Genes must be turned on or off at the right moments to ensure the proper progression of events.
Spatial Gene Expression: Spatial regulation determines where genes are expressed within an organism. Different cells and tissues have distinct roles, and precise gene expression localization is crucial for achieving cellular specialization and forming complex structures. This spatial control ensures that cells in different regions adopt specific identities and functions.
Signaling Pathways: Cellular signaling pathways play a crucial role in spatiotemporal gene expression. Signaling molecules transmit information from the extracellular environment to the cell's nucleus, where they influence gene transcription. This allows cells to interpret their surroundings and adjust gene expression accordingly.
Transcription Factors: Transcription factors are proteins that bind to specific DNA sequences and regulate gene expression. Their presence or absence can turn genes on or off, and their activity can be influenced by various factors, including signaling pathways and cellular context.
Epigenetic Modifications: Epigenetic modifications, such as DNA methylation and histone modifications, play a role in regulating gene expression patterns. These modifications can influence the accessibility of genes to transcription machinery, thereby impacting their activity.
Developmental Processes: During development, genes are expressed in specific patterns that guide the formation of tissues, organs, and body structures. This requires precise spatiotemporal coordination to ensure that genes are activated or repressed in the right places and at the right times.
Environmental Responses: Cells also adjust their gene expression patterns in response to environmental changes. This adaptability allows organisms to survive and thrive in different conditions by activating genes that provide a selective advantage.
Disease and Dysfunction: Dysregulation of spatiotemporal gene expression can lead to diseases, including developmental disorders and cancer. Understanding the underlying mechanisms of precise gene regulation is essential for deciphering disease origins and developing targeted therapies.
Orchestrating precise gene expression in time and space is a remarkable feat of biological complexity. This regulation enables organisms to navigate intricate developmental pathways, respond to changing environments, and maintain proper function. The interplay of signaling, transcription factors, epigenetic modifications, and other molecular processes ensures that genes are turned on or off with remarkable precision, ultimately shaping the remarkable diversity and functionality of life.
Mechanisms governing spatiotemporal gene patterns in diverse processes
The establishment and maintenance of precise spatiotemporal gene expression patterns are governed by a complex interplay of mechanisms that contribute to the diversity of biological processes. These mechanisms ensure that genes are activated or repressed at specific times and in specific locations, playing critical roles in development, adaptation, and cellular function.
Transcriptional Regulation: Transcription factors and enhancers are key players in spatiotemporal gene expression. Transcription factors bind to DNA sequences and activate or suppress gene transcription. Enhancers are DNA elements that can be far from the gene they regulate and interact with transcription factors to fine-tune gene expression in specific contexts.
Epigenetic Modifications: Epigenetic marks, such as DNA methylation and histone modifications, influence gene accessibility. These modifications can act as "marks" that determine whether a gene is actively transcribed or silenced, contributing to spatiotemporal regulation.
Chromatin Remodeling: Chromatin structure can be altered by chromatin remodeling complexes, making genes more or less accessible to transcription machinery. This dynamic alteration plays a role in controlling gene expression patterns.
Non-Coding RNAs: Non-coding RNAs, including microRNAs and long non-coding RNAs, can regulate gene expression by binding to target messenger RNAs (mRNAs) and affecting their stability or translation. They contribute to fine-tuning gene expression in specific tissues or developmental stages.
Cellular Signaling Pathways: Extracellular signals, transmitted through signaling pathways, influence gene expression. Activation of specific pathways in response to environmental cues or developmental signals can lead to changes in gene expression patterns.
Feedback Loops: Feedback loops involve regulatory proteins that control their own expression. These loops contribute to maintaining stable gene expression patterns and responding to fluctuations in cellular conditions.
Splicing and Alternative Promoters: Different exons can be included or excluded from mRNAs through alternative splicing, resulting in protein isoforms with distinct functions. Alternative promoters can also drive tissue-specific gene expression by producing different mRNA variants.
Nuclear Organization: The three-dimensional organization of the nucleus can influence gene expression. Genes located in close proximity within the nucleus may share regulatory elements, affecting their coordinated expression.
Cell-Cell Communication: Cells communicate with each other through signaling molecules that influence neighboring cells' gene expression patterns. This communication is crucial for coordinating development and maintaining tissue integrity.
Feedback and Feedforward Networks: Regulatory networks involving multiple genes can create feedback and feedforward loops, which contribute to robust and coordinated gene expression patterns.
The intricate interplay of these mechanisms allows organisms to generate a wide range of spatiotemporal gene expression patterns, facilitating processes such as embryonic development, tissue regeneration, immune responses, and adaptation to changing environments. The diversity of these mechanisms underscores the complexity of life's regulatory processes and highlights the remarkable precision with which genes are controlled to ensure optimal function.

AP patterning starts with maternal coordinate genes that regulate the expression of downstream zygotic genes in a hierarchic fashion. The maternal factors Bicoid and Nanos activate the expression of zygotic gap genes (hunchback, tailless, giant, Krüppel, knirps, and others). The products of the gap genes, combined with maternal morphogens, in turn, control the expression of pair-rule genes in seven stripes (e.g., even-skipped and fushi tarazu). Finally, the AP polarity of the parasegments is controlled by segment polarity genes, among which wingless (wg) and engrailed (en) are expressed along the segmental boundaries. 1
Deciphering the evolutionary timeline of spatiotemporal gene controls
The evolution of spatiotemporal gene controls would have unfolded over millions of years, driven by the gradual emergence of complex regulatory mechanisms that enabled organisms to adapt to diverse environments and developmental demands.
Early Gene Expression Patterns: In the earliest stages of life, simple organisms would have exhibited basic gene expression patterns that responded to fundamental environmental cues. These patterns would have been essential for survival and reproduction but lacked the intricate spatiotemporal specificity seen in more complex organisms.
Multicellular Evolution and Tissue Formation: With the emergence of multicellularity, the need for specialized cell types and tissues would have driven the development of more refined gene expression controls. Basic spatial patterns of gene expression would have been established to coordinate cell differentiation and contribute to the functionality of distinct cell populations.
Spatial Patterning in Development: As organisms diversified and developed more complex body plans, the establishment of precise spatial gene expression patterns would have become crucial. Regulatory mechanisms would have evolved to ensure that genes are activated or repressed in specific regions, contributing to the formation of intricate structures and organs.
Evolution of Signaling Pathways: The emergence of sophisticated signaling pathways and networks would have allowed organisms to respond to a wider range of environmental cues. These pathways would have interacted with gene regulatory elements, enabling cells to fine-tune their responses based on spatial and temporal inputs.
Cellular Differentiation and Specialization: As organisms evolved, more specialized cell types and tissues would have developed. This required the evolution of mechanisms that finely control gene expression to maintain cell identity and function within specific spatial contexts.
Fine-Tuning Developmental Timing: The evolution of spatiotemporal gene controls would also encompass the regulation of developmental timing. Genes would have been activated or repressed in specific sequences and at specific stages to ensure the proper progression of developmental events.
Adaptation and Environmental Responses: Organisms would have faced changing environments and selective pressures. The evolution of spatiotemporal gene controls would have allowed for the adaptation of gene expression patterns in response to new ecological niches, optimizing survival and reproduction.
Increased Complexity and Diversity: As evolution continued, organisms with more intricate and versatile spatiotemporal gene control mechanisms would have gained a competitive edge. This would have driven the diversification of developmental strategies and the emergence of complex life forms.
Co-Evolution of Mechanisms: The various mechanisms involved in spatiotemporal gene control, such as transcriptional regulation, epigenetic modifications, and cellular signaling, would have co-evolved to ensure their compatibility and effectiveness in coordinating gene expression.
The evolution of spatiotemporal gene controls represents a gradual process intertwined with the increasing complexity and diversity of life forms. These mechanisms would have played a pivotal role in shaping the development, adaptation, and functionality of organisms, contributing to the remarkable biological diversity observed in the natural world.
Genetic Components Essential for Spatiotemporal Gene Regulation
Spatiotemporal gene regulation is a complex process that involves the interplay of various genetic components, each contributing to the precise control of gene expression patterns in different regions and at different times within an organism. These components ensure the proper development, function, and adaptation of cells and tissues.
Promoters and Enhancers: Promoters are DNA sequences located near the start of a gene that serve as binding sites for RNA polymerase, initiating transcription. Enhancers are regulatory DNA elements that can be located far from the gene they regulate. They interact with transcription factors and other regulatory proteins to enhance or suppress gene expression in specific contexts.
Transcription Factors: Transcription factors are proteins that bind to DNA sequences and influence gene expression. They include activators that promote transcription and repressors that inhibit it. The presence and activity of specific transcription factors determine which genes are expressed and when.
Cis-Regulatory Modules: Cis-regulatory modules are combinations of enhancers and promoters that work together to regulate gene expression. These modules can contain multiple binding sites for different transcription factors, allowing for precise spatiotemporal control.
Epigenetic Marks: Epigenetic modifications, such as DNA methylation and histone modifications, influence gene accessibility. These marks can be inherited through cell divisions and contribute to the establishment of stable gene expression patterns.
Chromatin Remodeling Complexes: These complexes alter the structure of chromatin, making genes more or less accessible to transcription machinery. They play a role in fine-tuning gene expression patterns by controlling the availability of regulatory elements.
Non-Coding RNAs: Non-coding RNAs, including microRNAs and long non-coding RNAs, regulate gene expression by binding to target mRNAs. They can influence mRNA stability or translation efficiency, contributing to spatiotemporal regulation.
Cellular Signaling Pathways: Signaling pathways transmit information from the extracellular environment to the nucleus. They can activate or inhibit transcription factors, influencing gene expression based on developmental cues or environmental conditions.
Nuclear Organization: The three-dimensional organization of the nucleus can affect gene expression. Genes located in close proximity within the nucleus may share regulatory elements, allowing for coordinated expression.
Alternative Splicing: Many genes undergo alternative splicing, producing multiple mRNA isoforms from a single gene. This diversifies protein products and contributes to tissue-specific gene expression.
Feedback and Feedforward Loops: Regulatory networks involving multiple genes can create feedback and feedforward loops that contribute to robust and coordinated gene expression patterns.
Evolutionarily Conserved Elements: Some genetic elements are conserved across species and play critical roles in spatiotemporal gene regulation. These elements are often indicative of their importance in fundamental developmental processes.
Genome Accessibility Factors: Factors that influence chromatin accessibility, such as chromatin remodelers and pioneer factors, ensure that genes can be transcribed when needed.
Chromatin Boundary Elements: These elements define the boundaries of regulatory domains, preventing the spread of regulatory signals to inappropriate genes.
Adaptive Evolution of Regulatory Elements: As organisms adapt to new environments, the genetic components involved in spatiotemporal gene regulation can evolve to optimize gene expression for the organism's survival and fitness.
The coordination and interaction of these genetic components create a sophisticated regulatory network that enables precise spatiotemporal control of gene expression, allowing organisms to develop, function, and adapt in complex and dynamic environments.
Manufacturing codes steering spatiotemporal gene expression
The intricate process of spatiotemporal gene expression can be likened to the execution of manufacturing codes that guide the precise activation or repression of genes in specific spatial and temporal contexts. These "codes" involve a complex interplay of molecular components that work together to orchestrate the developmental and functional processes within an organism.
Genetic Blueprint: The DNA sequence itself serves as the foundational blueprint containing the instructions for building and regulating an organism. Specific sequences act as promoters, enhancers, and regulatory elements that are essential for controlling gene expression.
Transcription Factors as Programmers: Transcription factors can be thought of as programmers that read the genetic code and interpret it to initiate or suppress gene transcription. These proteins recognize specific DNA sequences and bind to them, initiating the transcriptional machinery.
Enhancers as Activation Codes: Enhancers can be likened to activation codes that, when bound by transcription factors, enhance gene expression. The presence of particular combinations of transcription factors at enhancers determines the level and timing of gene activation.
Repressors as Inhibitory Codes: Repressors function as inhibitory codes that prevent gene expression. They can bind to DNA sequences or interfere with the activity of transcription factors, ensuring that certain genes remain inactive in specific cellular contexts.
Epigenetic "Locks" and "Keys": Epigenetic modifications can be likened to locks and keys that regulate access to genes. Methylation and histone modifications serve as locks, rendering genes inaccessible. Conversely, specific transcription factors and co-activators act as keys that unlock genes for transcription.
Chromatin Remodeling as Assembly Lines: Chromatin remodeling complexes can be envisioned as assembly lines that modify the structure of chromatin, making genes more accessible or repressed. These complexes modify the chromatin landscape, allowing for the execution of gene expression codes.
Cell Signaling as Communication Protocols: Cellular signaling pathways function as communication protocols that relay external cues to the nucleus. They convey information about environmental conditions and developmental stages, guiding the execution of appropriate gene expression programs.
Non-Coding RNAs as Messengers: Non-coding RNAs act as messengers that transmit regulatory information to target genes. MicroRNAs and long non-coding RNAs can interact with mRNA molecules, influencing their stability and translation efficiency.
Evolution as Iterative Design Process: The evolutionary process can be seen as an iterative design process that refines the manufacturing codes over generations. Beneficial changes in regulatory elements and transcription factor binding sites are selected for, optimizing the precision and adaptability of gene expression.
Cellular Identity and Specialization as Output: The culmination of these manufacturing codes is the establishment of cellular identity and specialization. Cells "read" the codes to determine their roles and functions within the organism, contributing to the overall structure and functionality of tissues and organs.
Just as manufacturing codes ensure the proper assembly and functioning of complex machinery, the intricate genetic components, and mechanisms that govern spatiotemporal gene expression ensure the precise development, adaptation, and coordination of biological systems in the exquisite tapestry of life.
Epigenetic Tools Modulating Timely Gene Activations
Epigenetic mechanisms play a crucial role in regulating the timing of gene activations, contributing to the precise orchestration of gene expression patterns during development and in response to various environmental cues. These epigenetic tools provide a dynamic and adaptable framework for ensuring that genes are activated at the right time and in the right context.
DNA Methylation: DNA methylation involves the addition of methyl groups to cytosine bases, often leading to gene repression. During development, specific genes may be temporarily methylated, preventing their expression at inappropriate stages. Conversely, demethylation events can "unleash" genes when the time is right for their activation.
Histone Modifications: Histone modifications, such as acetylation, methylation, and phosphorylation, influence chromatin structure and gene accessibility. These modifications serve as epigenetic marks that can promote or inhibit transcription. Enzymes that add or remove these marks act as tools for regulating gene activation timing.
Chromatin Remodeling Complexes: Chromatin remodeling complexes utilize energy to reposition, evict, or alter nucleosomes, affecting the accessibility of genes to transcription machinery. These complexes act as tools for modifying chromatin structure and allowing genes to be activated or repressed when needed.
Long Non-Coding RNAs (lncRNAs): Long non-coding RNAs can interact with chromatin-modifying complexes and serve as guides for their recruitment to specific genomic regions. They act as epigenetic tools that contribute to the establishment of proper gene expression patterns during development.
Polycomb Group Proteins: Polycomb group proteins form complexes that regulate the maintenance of gene silencing during development. These proteins serve as epigenetic tools that ensure certain genes remain inactive until the appropriate developmental stage, preventing premature activation.
Chromatin States and Bivalent Domains: Certain genomic regions exhibit "bivalent" domains that possess both activating and repressive histone marks. These domains keep critical developmental genes poised for activation, allowing for rapid responses to cues while maintaining repression until needed.
Developmental Clocks and Timers: Epigenetic changes can be linked to internal biological clocks that regulate gene expression timing. Circadian rhythms, for instance, involve epigenetic modifications that enable genes to be activated or repressed according to specific time windows.
Environmental Sensing and Adaptation: Epigenetic modifications can be influenced by environmental factors, allowing organisms to adapt their gene expression patterns in response to changing conditions. This epigenetic plasticity acts as a tool for timely adjustments to various stimuli.
Evolutionary Conservation of Epigenetic Mechanisms: The conservation of epigenetic tools across species suggests their importance in regulating timely gene activations. These mechanisms have been refined over evolutionary time to optimize the timing of gene expression for the development and survival of organisms.
Overall, epigenetic tools serve as intricate mechanisms for modulating timely gene activations. They allow organisms to fine-tune their gene expression patterns in response to developmental cues, environmental changes, and physiological demands, ensuring that genes are activated or repressed at the appropriate moments for optimal function.
Interplay between Signaling Pathways and Gene Expression Patterns
The interplay between signaling pathways and gene expression patterns is a dynamic and intricate process that governs how cells and organisms respond to their environment, regulate development, and maintain homeostasis. This coordination ensures that cellular activities are precisely controlled and adapted to changing conditions.
Sensory Perception and Signal Initiation: Signaling pathways are initiated by sensory inputs, such as growth factors, hormones, or environmental cues. These signals are detected by receptors on the cell surface or within the cell, triggering a cascade of events.
Transduction and Signal Amplification: Once a signal is detected, it is transduced into a series of biochemical events within the cell. This often involves amplification of the signal, where a single extracellular ligand can lead to the activation of multiple intracellular molecules.
Activation of Transcription Factors: Signaling pathways frequently converge on transcription factors. Activated transcription factors are then transported to the nucleus, where they bind to specific DNA sequences, promoting or repressing gene transcription.
Gene Expression Changes: The binding of transcription factors to DNA directly influences gene expression. Genes targeted by transcription factors associated with a specific signaling pathway will be activated or repressed based on the pathway's activation status.
Fine-Tuning Gene Regulation: Multiple signaling pathways can converge on a single gene, providing layers of regulation. This fine-tuning ensures that gene expression is responsive to a combination of signals, allowing for more precise control.
Feedback Loops: Signaling pathways can regulate their own activity through feedback loops. Genes activated by a signaling pathway can encode proteins that modulate the pathway itself, leading to self-regulation.
Cross-Talk between Pathways: Signaling pathways can cross-talk, where components of one pathway influence the activity of another. This integration allows cells to integrate multiple inputs and generate a coordinated response.
Developmental Patterning: Signaling pathways are instrumental in establishing developmental patterns. During embryogenesis, for instance, gradients of signaling molecules can lead to distinct gene expression patterns that define tissue identities.
Environmental Adaptation: Signaling pathways enable cells to respond to changes in their environment. Cells can adjust their gene expression patterns to cope with varying conditions, optimizing their survival and function.
Dysregulation and Disease: Dysregulation of signaling pathways can lead to diseases. Aberrant activation of oncogenic signaling pathways can result in uncontrolled cell growth and cancer. Similarly, improper signaling can contribute to developmental disorders.
Evolutionary Adaptation: Signaling pathways have evolved to allow organisms to adapt to different environments and challenges. Evolutionary changes in signaling components can lead to modified gene expression patterns that confer selective advantages.
The intricate interplay between signaling pathways and gene expression patterns forms a complex regulatory network that ensures cells respond appropriately to their surroundings, guiding their development, function, and adaptation. This interwoven system highlights the remarkable coordination required for cells and organisms to thrive in diverse and ever-changing environments.
Regulatory Systems Harmonizing Spatiotemporal Gene Expression
The harmonization of spatiotemporal gene expression is orchestrated by a sophisticated network of regulatory systems that work in concert to ensure precise and coordinated gene activation or repression in specific spatial and temporal contexts. This orchestration is fundamental for proper development, functionality, and adaptability within complex biological systems.
Transcriptional Regulation and Enhancer-Promoter Interactions: Transcription factors and enhancers play a pivotal role in coordinating spatiotemporal gene expression. Transcription factors bind to enhancers and promoters, facilitating communication between distal regulatory elements and gene promoters, ensuring genes are activated or repressed when and where needed.
Epigenetic Mechanisms and Chromatin Remodeling: Epigenetic modifications and chromatin remodeling contribute to the establishment of permissive or repressive chromatin states. These mechanisms enable genes to be accessible or inaccessible to the transcription machinery, fine-tuning gene expression patterns across different cellular contexts.
Cellular Signaling Pathways and Signal Integration: Cellular signaling pathways converge on transcription factors, influencing their activity based on external cues. Integration of multiple signaling inputs ensures that genes are activated or repressed in response to a combination of signals, leading to a coordinated cellular response.
Temporal Regulation and Developmental Clocks: Biological clocks and developmental timers dictate the timing of gene expression. These internal timing mechanisms synchronize with external cues to activate or repress genes at specific developmental stages or circadian rhythms.
Spatial Patterning and Morphogen Gradients: Morphogen gradients establish spatial patterns of gene expression during embryogenesis. The concentration of signaling molecules provides positional information that guides the activation or repression of genes, contributing to the formation of distinct tissue identities.
Feedback and Feedforward Loops: Regulatory networks often incorporate feedback and feedforward loops. These loops maintain stable gene expression patterns and enable rapid responses to changes in cellular conditions, ensuring robustness and precision.
Cell-Cell Communication and Signaling Crosstalk: Cells communicate and coordinate their gene expression through signaling molecules. Signaling crosstalk allows neighboring cells to influence each other's gene expression patterns, contributing to the organization and function of tissues.
Alternative Splicing and Isoform Diversity: Alternative splicing generates different mRNA isoforms from a single gene, resulting in protein diversity. This mechanism contributes to the specialization of cell types and tissues through the production of distinct protein isoforms.
Evolutionary Conservation and Adaptation: Regulatory systems controlling spatiotemporal gene expression are evolutionarily conserved across species. While core mechanisms remain similar, variations allow organisms to adapt and optimize gene expression for their specific needs and environments.
Multilevel Integration and Robustness: The integration of various regulatory systems ensures the robustness of spatiotemporal gene expression. This multilevel coordination minimizes errors and deviations, maintaining the fidelity of developmental and physiological processes.
The intricate interplay of these regulatory systems creates a symphony of gene expression that harmonizes across different spatial and temporal scales, culminating in the intricate diversity and functionality observed within living organisms. This precise orchestration is essential for shaping the complexity of life and enabling organisms to thrive in diverse ecological niches.
Is there evidence for evolutionary mechanisms instantiating intricate gene expression controls?
Gene expression is an immensely intricate process. Every step in this process, from DNA transcription to protein synthesis, requires specific codes, signaling pathways, and molecular machinery. For the machinery to function properly, each of its many components must be precisely coordinated, leading many to question how such complexity could arise step-by-step through evolution. Consider the process of transcription, wherein a specific segment of DNA is read by RNA polymerase and transcribed into a complementary mRNA strand. This requires specific promoter regions on the DNA to signal the start of transcription and various transcription factors that recognize and bind to these promoters. The mere presence of these promoter regions and the specific binding proteins suggests an inherent coordination. Without these transcription factors, the promoters are useless. Conversely, without the promoters, the transcription factors have nothing to bind to. It's difficult to envision how one could evolve without the other, as each would bear no function on its own. Furthermore, the genetic code itself is a marvel of complexity. Each three-nucleotide codon on mRNA specifies a particular amino acid, a language that is then read and translated by ribosomes. This decoding process requires the precise interaction of mRNA with tRNA molecules that carry the appropriate amino acid. For this to function, not only does the mRNA codon need to exist and be transcribed correctly, but there also needs to be a corresponding tRNA molecule with the correct anticodon and attached amino acid, as well as the enzyme to attach that amino acid to the tRNA. The simultaneous existence and functional integration of all these parts seem improbable to have evolved in isolation. Then, there's the post-translational modification of proteins. Many proteins, once synthesized, undergo modifications like phosphorylation, methylation, or glycosylation. These modifications are essential for the protein's function, stability, or localization. The enzymes that carry out these modifications must recognize specific motifs on the proteins. Again, this implies a coordinated existence. The motif on the protein is pointless without the modifying enzyme, and the enzyme has no function without the protein motif.
Lastly, cellular signaling pathways show a level of interdependence and specificity that's breathtaking. For a signal to be transduced from the cell membrane to the nucleus, or between cells, numerous proteins and molecules need to interact in a highly regulated sequence. Missing just one component or having one non-functional component can disrupt the entire pathway. Given the intricacies of gene expression, it's hard to imagine how the multitude of processes and their respective molecular players could have evolved piecemeal. They seem to necessitate a complete, fully operational system from the onset to function efficiently and with purpose. Without this full system in place, it seems unlikely that partial or transitional stages would confer any evolutionary advantage, leading many to propose that such complexity and interdependence may suggest the work of an intelligent architect.
Investigating spatiotemporal gene controls for irreducibility traits
The complexity and precision observed in the realm of spatiotemporal gene controls present fascinating insights into the depth of coordination required for cellular operations. These processes can be described as complex mechanisms that could be viewed as both irreducible and interdependent.
Transcriptional Machinery: This includes the DNA itself, RNA polymerase, transcription factors, and promoter regions on DNA. Without each of these components interacting in harmony, the transcription of DNA into mRNA would be impossible.
Genetic Code: The codons of mRNA, the anticodons of tRNA, and the amino acids they attach to all have to match perfectly. A mistake here would result in the wrong protein being synthesized.
Ribosomal Translation: Ribosomes, along with mRNA and tRNA, translate the genetic code into proteins. Without the precise interaction between these molecules, protein synthesis is halted.
Post-translational Modifications: Many proteins require modifications after being synthesized. These modifications often require specific enzymes that recognize certain motifs on the proteins. The motifs and the enzymes are useless without each other.
Cellular Signaling: From ligands binding to receptors on the cell membrane, to secondary messengers, to protein kinases, every component in a signaling pathway is essential. Without one component, the signal cannot be relayed.
DNA Replication and Repair: DNA polymerases, helicases, topoisomerases, and various repair enzymes ensure DNA is replicated and any errors are fixed. A malfunction in one could be catastrophic for the cell.
Epigenetic Regulation: Histone modifications, DNA methylation, and non-coding RNAs all play roles in determining when and how genes are expressed. The language they 'speak' ensures that genes are expressed at the right time and in the right cells.
All these systems exhibit a form of irreducibility and interdependence. For example, in the case of the transcriptional machinery, transcription factors need promoters to bind to, while the promoters require the transcription factors for recognition. In terms of cellular signaling, a ligand without a receptor, or a kinase without a substrate, is functionally redundant. The codes and languages governing these processes crosstalk and integrate. DNA methylation might guide transcription factors, and ligands might trigger a cascade affecting gene expression. Given the level of coordination and the interplay between these systems, it's challenging to understand how such processes could evolve in isolation. The half-formed or transitional stages of such systems might not confer any evolutionary advantage, suggesting that such intricate systems, codes, and languages would need to be in place from the outset for efficient and purposeful function. The existence of such coordinated complexity may lead some to believe that they are the outcome of deliberate design.
Coordinated interactions post-establishment of spatiotemporal gene systems
The establishment of spatiotemporal gene systems ensures that genes are expressed in the right cells, at the right time, and in response to the right stimuli. But the complexity doesn't end there. Post-establishment of these gene systems, an intricate array of coordinated interactions is essential to maintain, modulate, and respond to the cellular environment. These interactions, functioning as a symphony of molecular events, are foundational to the robust and dynamic nature of cellular life.
Signal Transduction: Cells are in constant dialogue with their surroundings. External signals, whether they be hormones, nutrients, or other molecules, bind to cell receptors, triggering a cascade of intracellular events. These pathways often converge on transcriptional regulators, modifying gene expression in response to external cues.
Feedback Loops
Negative Feedback: Often, the product of a gene's expression will inhibit further expression of that gene. This ensures that cells don't overproduce certain proteins and maintain homeostasis.
Positive Feedback: Conversely, in some cases, the product of a gene might enhance its further expression. This can be crucial for rapid responses, such as in immune activation.
Tissue-specific Gene Regulation
Cell-to-Cell Communication: Cells within tissues don't act in isolation. Through mechanisms like paracrine signaling, one cell can influence the gene expression of its neighbors.
Epigenetic Landscaping: Epigenetic markers, influenced by both internal cellular states and external factors, play a significant role in tissue-specific gene expression. As cells differentiate, certain genes are 'locked' into on or off states, ensuring that cells maintain their specific identities.
Integration with Metabolic Pathways
Metabolite Regulation: Metabolic byproducts can directly influence gene expression. For example, when energy is abundant, high levels of ATP can impact the genes related to energy storage.
Temporal Regulation and Circadian Rhythms
Circadian Clocks: Many organisms have evolved internal clocks that regulate gene expression in 24-hour cycles. This ensures that certain processes, like DNA repair or photosynthesis, occur at optimal times of the day.
These multifaceted interactions underscore a profound interconnectedness within cellular systems. The regulation and coordination don't merely rest on establishing the spatiotemporal gene systems but persist throughout the cell's lifecycle. The exquisite precision and synchronized dance of these molecular interactions showcase the depth of complexity within even the simplest of cells. This might lead some to marvel at the ingenuity of such a system, wondering if such intricate coordination is the product of a specific design.
1. All systems that rely on semiotic code, languages, are interdependent and must emerge together interlocked, exhibit characteristics of deliberate design.
2. The processes of gene expression, transcription, translation, and post-translational modifications rely on semiotic code, languages, are interdependent, and must emerge together interlocked.
3. Therefore, the processes of gene expression, transcription, translation, and post-translational modifications exhibit characteristics of deliberate design.
1. Gheisari, E., Aakhte, M., & Müller, H.-A. J. (2020). Gastrulation in Drosophila melanogaster: Genetic control, cellular basis and biomechanics. Mechanisms of Development, 163, 103629. https://doi.org/10.1016/j.mod.2020.103629


