Microorganisms contributing to the Nitrogen Cycle
Two kinds of nitrogen-fixing bacteria are recognized. The first kind, the free-living (nonsymbiotic) bacteria. The second kind comprises the mutualistic (symbiotic) bacteria; examples include Rhizobium, associated with leguminous plants
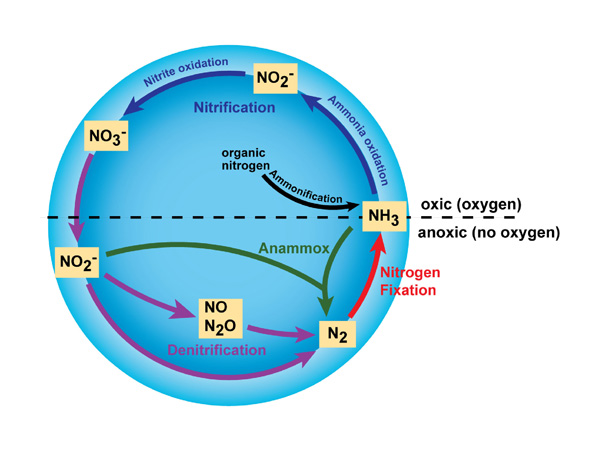
Denitrification
Denitrifying bacteria have been identified in over 50 genera with over 125 different species and are estimated to represent 10-15% of bacteria population in water, soil and sediment2
Paracoccus denitrificans
Pseudomonas stutzeri
Pyrobaculum aerophilum
Haloarcula marismortui
Fusarium oxysporum
Methanosarcinales
Shewanella baltica
Few out of the over 100 currently identifiable major lineages or phyla within the domain Bacteria have nitrogen-fixing members, namely
Nitrogen-fixing prokaryotes
Actinobacteria
Acidobacteria
Alphaproteobacteria
Azoarcus
Azotobacteraceae
Azospirillum
Azotobacter vinelandii
Acetobacter
Bacteroidetes
Beijerinckia
Bradyrhizobium
Capparis
Crenarchaeota
Cyanobacteria Calvin–Benson cycle
Chlorobi Reductive Tricarboxylic Acid Cycle
Clostridium
Cyanothece
Deschampsia caespitosa
Derxia
Enterobacter
Erwinia herbicola
Euryarchaeota
Frankia
Gammaproteobacteria
Gluconacetobacter diazotrophicus
Gluconacetobacter,
Gluconobacter
Herbaspirillum lusitanum
Klebsiella variicola
Klebsiella pneumoniae
Methylacidiphilum fumariolicum
Mesorhizobium
Mesorhizobium
Ochrobactrum
Parvarchaeota
Pantoea
Proteobacteria
Planctomycetes
Paenibacillus
Pseudomonas stutzeri
Rhodospirillaceae
Rhizobiales
Rhizobium
Sinorhizobium
Spirillum
Stenotrophomonas
Trichodesmium
Verrucomicrobia
Verrucomicrobia
The symbiotic N2-fixation in angiosperms
Symbiotic associations occur in every habitat on earth, but we know very little about their evolutionary histories. 1 Traits arising from symbiotic partnerships require complex coordination between different species. This type of complex coordination is especially evident in the symbiosis between angiosperms and their N2-fixing bacterial symbionts. Although progress has been made in cataloging the distinct morphologies and diverse microbial partnerships characterizing N2-fixing symbioses and in the molecular mechanisms regulating the symbiosis in some plant species, it is unknown how all this novelty was generated. Such diversity and wide taxonomic distribution could suggest multiple, independent regulatory circuits that arose de novo. If there is no deep symbiotic homology, there are potentially multiple pathways towards achieving nodulation.
My comment: What: If de novo, no evolutionary tree then? And if there are multiple pathways, the author entirely ignores and misses to ask, what mechanisms could account for those de novo pathways.
Our model unambiguously pinpoints the single origin of the precursor within the Fabidae (also known as Rosids I) at the base of the four orders comprising the N2-fixing clade. Although there is one single precursor origin, we found ~8 subsequent origins of N2-fixation itself leading to distinct symbiosis types in the angiosperms (most notably those involving rhizobial bacteria and those with actinorhizal Frankia sp.). The dramatic differences in infection modes, nodule forms, symbiont identity, and resource control processes among N2 fixers6 demonstrate that substantial diversification of nodulation types took place following the origin of the shared precursor.
It has been suggested that abiotic sources of fixed nitrogen on the early Earth, supplied through endogenous/abiotic synthesis and exogenous delivery, were most likely limiting.
My comment: Why do the authors not specify what sources these would have been?
At some point, dwindling concentrations of reduced nitrogen would have become insufficient for an expanding microbial biomass, precipitating the evolution of biological nitrogen fixation. These prevailing conditions have been used to argue that the innovation of biological nitrogen fixation occurred early in prokaryotic evolution, and indeed the ability to fix nitrogen is found exclusively among members of the bacteria and archaea. Maintaining the ability to fix nitrogen in the presence of exogenous or endogenous sources of O2 has necessitated innovative biochemical and physiological mechanisms for segregation. This is particularly important in oxygen-evolving cyanobacteria and organisms carrying out oxidative phosphorylation. Certain cyanobacteria contribute substantial amounts of fixed nitrogen in marine environments and do so because of exquisite controls on temporal and spatial separation of the two processes
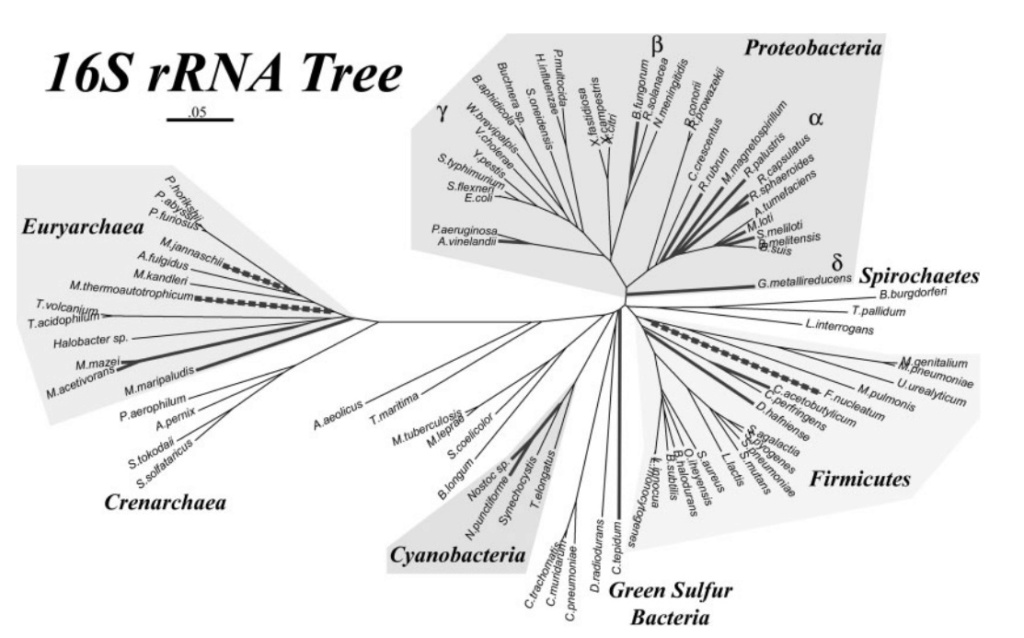
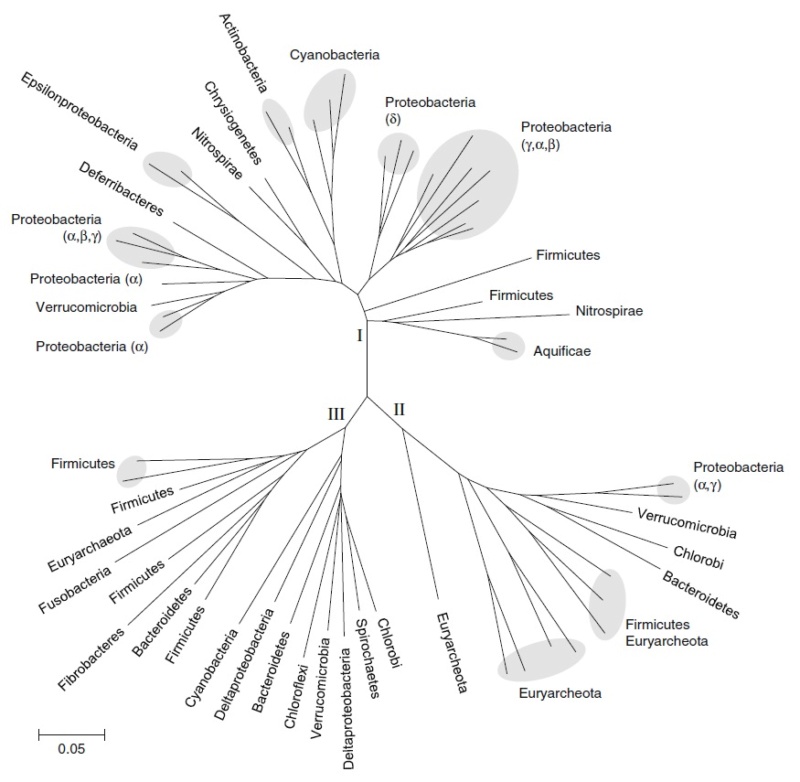
Neighbor-joining phylogenetic tree constructed from 16S rDNA sequences for the microbial genomes used in this analysis. Diazotrophic genomes, as determined by the presence of NifHDKEN operons, are indicated with bold lines. Lineages outlined by dashed bold lines have homologs to NifH and NifD but are not known to fix nitrogen. Also shown are the major bacterial and archaeal phyla, highlighted if nitrogen-fixing lineages are found among them
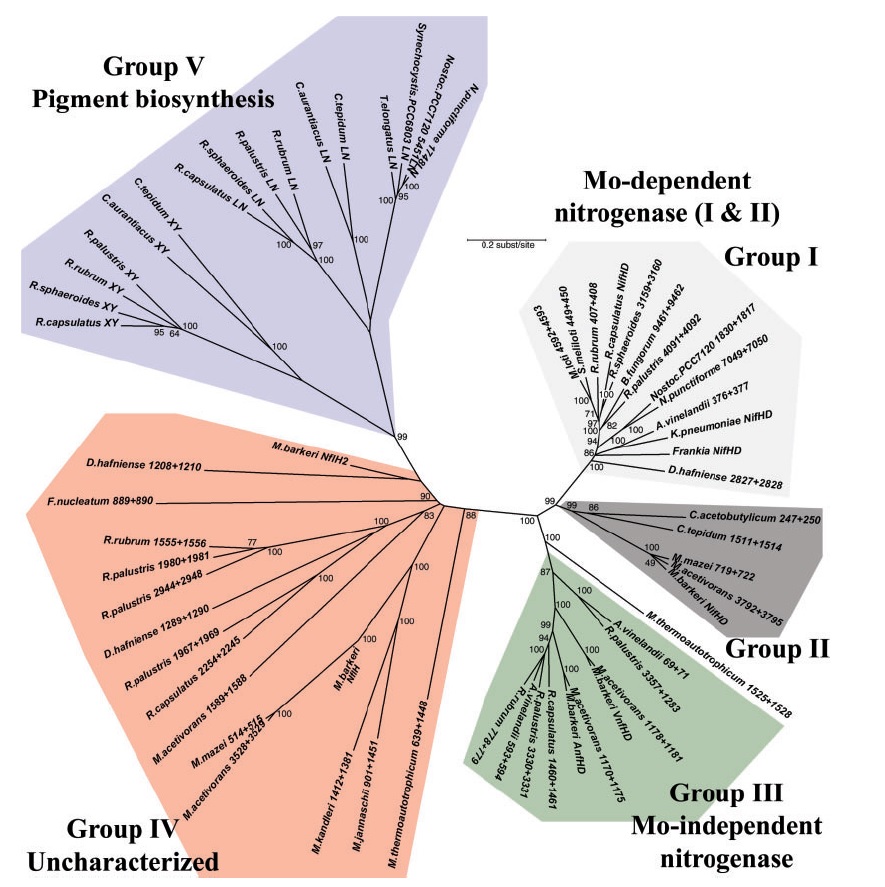
Overview of five phylogenetic groups elucidated in the text, shown on a concatenated phylogenetic tree composed of NifH and NifD homologs found in complete genomes. Groups I to III are all functional nitrogenases, including Mo-dependent group I and II nitrogenases as well as vanadium-dependent and iron-dependent group III homologs. Group V includes the subunits of protochlorophyllide reductase (here, BchX and Y concatenated) and chlorophyllide reductase (BchL and N concatenated) involved in the crucial late steps in photosynthetic pigment biosynthesis, which segregate into two distinct clades. Group IV consists of yet to be characterized nitrogenase paralogs of the NifH and NifD proteins, many of which are present in organisms not known to fix nitrogen. Because of significant sequence divergence, group IV homologs do not resolve to a single origin and so the branching within this group should be interpreted cautiously.

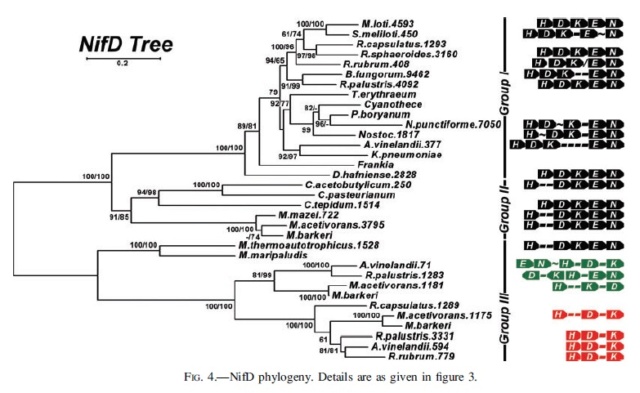
NifH phylogeny. Shown at the right of each figure is the position of the nitrogenase groups outlined in the text, as well as the local nitrogenase operon structure showing the relative positions of the HDKEN proteins (operons are color coded in black, green, or red for Mo-dependence, V-dependence, or Fe-dependence, respectively). For organisms with whole genome sequences available, the number after the organism name indicates the relative position of the protein’s open reading frame within that genome (except in the cases of R. capsulatus and R. palustris). Within the operon structures, each dash (-) indicates a single open reading frame, the approximation symbol (; ) denotes five to 10 open reading frames, and the slash (/) indicates that there is no clear local/operonal association between genes. Numbers at tree nodes indicate bootstrap support based on 100 maximum-likelihood replicates (first number) or 500 distance replicates (second number). The background tree is the distance tree; a short dash (-) at a node indicates low bootstrap support (,60%) for that branching pattern for either the ML tree or distance tree (although the same topology was observed). A single number or no number indicates a disagreement in the topology at that node between the ML and distance trees, with a single number indicating bootstrap support from the distance tree if it is 60% or greater. The scale bar gives the number of substitutions per site for branches on the distance tree.

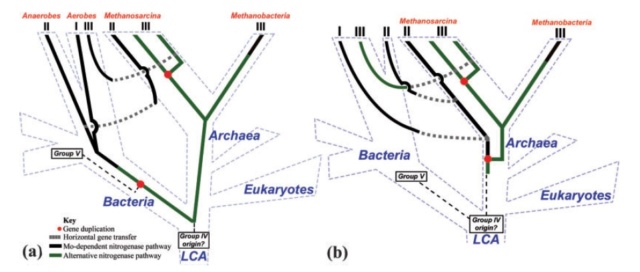
Proposed gene versus species trees, as discussed in the text. (a) Nitrogen-fixing LCA hypothesis, showing the three domains and their divergence from the LCA in blue text and dashed blue lines. Solid lines (black and green, depending on hypothesized metal specificity) indicate the evolution of nitrogenase from the base of the tree a group IV ancestor. Also indicated are putative gene duplication (red dots) and horizontal gene transfer (originating at gray boxes, with transfer indicated by gray dashed lines) events. The three nitrogenase phylogenetic groups described in the text are indicated at the tips of the tree, as are the predominant organisms in which they are found. According to the LCA model, gene loss has been extensive and accounts for the majority of modern lineages not being able to fix their own nitrogen. (b) Methanogen origin hypothesis, using the same color scheme and symbols as figure 8a. According to this model, nitrogen fixation was invented in methanogenic archaea and subsequently was transferred into a primitive bacterium, circumventing the necessity for extensive gene loss to explain the paucity of diazotrophic lineages. As with the LCA hypothesis, several relatively recent HGT events must have occurred to explain the distribution and high identities of group II and III nitrogenases.
1. https://www.nature.com/articles/ncomms5087
2. https://en.wikipedia.org/wiki/Denitrifying_bacteria
Two kinds of nitrogen-fixing bacteria are recognized. The first kind, the free-living (nonsymbiotic) bacteria. The second kind comprises the mutualistic (symbiotic) bacteria; examples include Rhizobium, associated with leguminous plants

Denitrification
Denitrifying bacteria have been identified in over 50 genera with over 125 different species and are estimated to represent 10-15% of bacteria population in water, soil and sediment2
Paracoccus denitrificans
Pseudomonas stutzeri
Pyrobaculum aerophilum
Haloarcula marismortui
Fusarium oxysporum
Methanosarcinales
Shewanella baltica
Few out of the over 100 currently identifiable major lineages or phyla within the domain Bacteria have nitrogen-fixing members, namely
Nitrogen-fixing prokaryotes
Actinobacteria
Acidobacteria
Alphaproteobacteria
Azoarcus
Azotobacteraceae
Azospirillum
Azotobacter vinelandii
Acetobacter
Bacteroidetes
Beijerinckia
Bradyrhizobium
Capparis
Crenarchaeota
Cyanobacteria Calvin–Benson cycle
Chlorobi Reductive Tricarboxylic Acid Cycle
Clostridium
Cyanothece
Deschampsia caespitosa
Derxia
Enterobacter
Erwinia herbicola
Euryarchaeota
Frankia
Gammaproteobacteria
Gluconacetobacter diazotrophicus
Gluconacetobacter,
Gluconobacter
Herbaspirillum lusitanum
Klebsiella variicola
Klebsiella pneumoniae
Methylacidiphilum fumariolicum
Mesorhizobium
Mesorhizobium
Ochrobactrum
Parvarchaeota
Pantoea
Proteobacteria
Planctomycetes
Paenibacillus
Pseudomonas stutzeri
Rhodospirillaceae
Rhizobiales
Rhizobium
Sinorhizobium
Spirillum
Stenotrophomonas
Trichodesmium
Verrucomicrobia
Verrucomicrobia

The symbiotic N2-fixation in angiosperms
Symbiotic associations occur in every habitat on earth, but we know very little about their evolutionary histories. 1 Traits arising from symbiotic partnerships require complex coordination between different species. This type of complex coordination is especially evident in the symbiosis between angiosperms and their N2-fixing bacterial symbionts. Although progress has been made in cataloging the distinct morphologies and diverse microbial partnerships characterizing N2-fixing symbioses and in the molecular mechanisms regulating the symbiosis in some plant species, it is unknown how all this novelty was generated. Such diversity and wide taxonomic distribution could suggest multiple, independent regulatory circuits that arose de novo. If there is no deep symbiotic homology, there are potentially multiple pathways towards achieving nodulation.
My comment: What: If de novo, no evolutionary tree then? And if there are multiple pathways, the author entirely ignores and misses to ask, what mechanisms could account for those de novo pathways.
Our model unambiguously pinpoints the single origin of the precursor within the Fabidae (also known as Rosids I) at the base of the four orders comprising the N2-fixing clade. Although there is one single precursor origin, we found ~8 subsequent origins of N2-fixation itself leading to distinct symbiosis types in the angiosperms (most notably those involving rhizobial bacteria and those with actinorhizal Frankia sp.). The dramatic differences in infection modes, nodule forms, symbiont identity, and resource control processes among N2 fixers6 demonstrate that substantial diversification of nodulation types took place following the origin of the shared precursor.
It has been suggested that abiotic sources of fixed nitrogen on the early Earth, supplied through endogenous/abiotic synthesis and exogenous delivery, were most likely limiting.
My comment: Why do the authors not specify what sources these would have been?
At some point, dwindling concentrations of reduced nitrogen would have become insufficient for an expanding microbial biomass, precipitating the evolution of biological nitrogen fixation. These prevailing conditions have been used to argue that the innovation of biological nitrogen fixation occurred early in prokaryotic evolution, and indeed the ability to fix nitrogen is found exclusively among members of the bacteria and archaea. Maintaining the ability to fix nitrogen in the presence of exogenous or endogenous sources of O2 has necessitated innovative biochemical and physiological mechanisms for segregation. This is particularly important in oxygen-evolving cyanobacteria and organisms carrying out oxidative phosphorylation. Certain cyanobacteria contribute substantial amounts of fixed nitrogen in marine environments and do so because of exquisite controls on temporal and spatial separation of the two processes

Neighbor-joining phylogenetic tree constructed from 16S rDNA sequences for the microbial genomes used in this analysis. Diazotrophic genomes, as determined by the presence of NifHDKEN operons, are indicated with bold lines. Lineages outlined by dashed bold lines have homologs to NifH and NifD but are not known to fix nitrogen. Also shown are the major bacterial and archaeal phyla, highlighted if nitrogen-fixing lineages are found among them

Overview of five phylogenetic groups elucidated in the text, shown on a concatenated phylogenetic tree composed of NifH and NifD homologs found in complete genomes. Groups I to III are all functional nitrogenases, including Mo-dependent group I and II nitrogenases as well as vanadium-dependent and iron-dependent group III homologs. Group V includes the subunits of protochlorophyllide reductase (here, BchX and Y concatenated) and chlorophyllide reductase (BchL and N concatenated) involved in the crucial late steps in photosynthetic pigment biosynthesis, which segregate into two distinct clades. Group IV consists of yet to be characterized nitrogenase paralogs of the NifH and NifD proteins, many of which are present in organisms not known to fix nitrogen. Because of significant sequence divergence, group IV homologs do not resolve to a single origin and so the branching within this group should be interpreted cautiously.

NifH phylogeny. Shown at the right of each figure is the position of the nitrogenase groups outlined in the text, as well as the local nitrogenase operon structure showing the relative positions of the HDKEN proteins (operons are color coded in black, green, or red for Mo-dependence, V-dependence, or Fe-dependence, respectively). For organisms with whole genome sequences available, the number after the organism name indicates the relative position of the protein’s open reading frame within that genome (except in the cases of R. capsulatus and R. palustris). Within the operon structures, each dash (-) indicates a single open reading frame, the approximation symbol (; ) denotes five to 10 open reading frames, and the slash (/) indicates that there is no clear local/operonal association between genes. Numbers at tree nodes indicate bootstrap support based on 100 maximum-likelihood replicates (first number) or 500 distance replicates (second number). The background tree is the distance tree; a short dash (-) at a node indicates low bootstrap support (,60%) for that branching pattern for either the ML tree or distance tree (although the same topology was observed). A single number or no number indicates a disagreement in the topology at that node between the ML and distance trees, with a single number indicating bootstrap support from the distance tree if it is 60% or greater. The scale bar gives the number of substitutions per site for branches on the distance tree.



Proposed gene versus species trees, as discussed in the text. (a) Nitrogen-fixing LCA hypothesis, showing the three domains and their divergence from the LCA in blue text and dashed blue lines. Solid lines (black and green, depending on hypothesized metal specificity) indicate the evolution of nitrogenase from the base of the tree a group IV ancestor. Also indicated are putative gene duplication (red dots) and horizontal gene transfer (originating at gray boxes, with transfer indicated by gray dashed lines) events. The three nitrogenase phylogenetic groups described in the text are indicated at the tips of the tree, as are the predominant organisms in which they are found. According to the LCA model, gene loss has been extensive and accounts for the majority of modern lineages not being able to fix their own nitrogen. (b) Methanogen origin hypothesis, using the same color scheme and symbols as figure 8a. According to this model, nitrogen fixation was invented in methanogenic archaea and subsequently was transferred into a primitive bacterium, circumventing the necessity for extensive gene loss to explain the paucity of diazotrophic lineages. As with the LCA hypothesis, several relatively recent HGT events must have occurred to explain the distribution and high identities of group II and III nitrogenases.
1. https://www.nature.com/articles/ncomms5087
2. https://en.wikipedia.org/wiki/Denitrifying_bacteria

