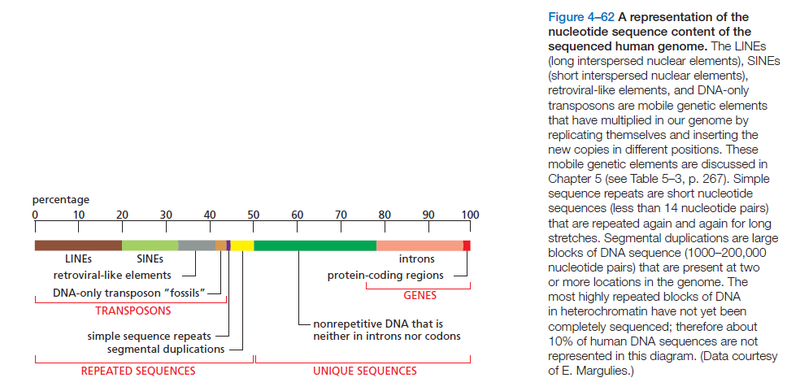
https://reasonandscience.catsboard.com/t2393-transposons-and-retrotransposons
Class I TEs are copied in two stages: first, they are transcribed from DNA to RNA, and the RNA produced is then reverse transcribed to DNA. This copied DNA is then inserted back into the genome at a new position. The reverse transcription step is catalyzed by a reverse transcriptase, which is often encoded by the TE itself. The characteristics of retrotransposons are similar to retroviruses, such as HIV.
Retrotransposons are commonly grouped into three main orders:
- TEs with long terminal repeats (LTRs), which encode reverse transcriptase, similar to retroviruses
- Long interspersed nuclear elements (LINEs, LINE-1s, or L1s), which encode reverse transcriptase but lack LTRs, and are transcribed by RNA polymerase II
- Short interspersed nuclear elements do not encode reverse transcriptase and are transcribed by RNA polymerase III
Class II (DNA transposons)
The cut-and-paste transposition mechanism of class II TEs does not involve an RNA intermediate. The transpositions are catalyzed by several transposase enzymes. Some transposases non-specifically bind to any target site in DNA, whereas others bind to specific target sequences. The transposase makes a staggered cut at the target site resulting in single-strand 5' or 3' DNA overhangs, so-called "sticky ends". This step cuts out the DNA transposon, which is then ligated into a new target site; the process involves activity of a DNA polymerase that fills in gaps and of a DNA ligase that closes the sugar-phosphate backbone.[citation needed] This results in duplication of the target site. The insertion sites of DNA transposons may be identified by short direct repeats (created by the staggered cut in the target DNA and filling in by DNA polymerase) followed by a series of inverted repeats important for the TE excision by transposase. Cut-and-paste TEs may be duplicated if their transposition takes place during S phase of the cell cycle, when a donor site has already been replicated but a target site has not yet been replicated.[citation needed] Such duplications at the target site can result in gene duplication, which plays an important role in genomic evolution.[14]:284 Not all DNA transposons transpose through the cut-and-paste mechanism. In some cases, a replicative transposition is observed in which a transposon replicates itself to a new target site (e.g. helitron (biology)).Class II TEs comprise less than 2% of the human genome, making the rest Class I.[15]
Why repetitive DNA is essential to genome function 1
The discovery of repetitive DNA presents a conceptual problem for traditional genebased notions of hereditary information. repetitive DNA is an essential component of genomes; it is required for formatting coding information so that it can be accurately expressed and for formatting DNA molecules for transmission to new generations of cells. In addition, the cooperative nature of proteinDNA interactions provides another fundamental reason why repeated sequence elements are essential to format genomic DNA. Instead of parasites, we argue that repetitive DNA elements are necessary organisers of genomic information.
molecular genetics has shown that achieving this task requires cells to possess a number of additional capabilities also encoded in the genome:
(1) Regulating timing and extent of coding sequence expression.
(2) Organizing coordinated expression of protein and RNA molecules that function together.
(3) Packaging DNA appropriately within the cell.
(4) Replicating the genome in synchrony with the cell division cycle.
(5) Transmitting replicated DNA accurately to progeny cells at cell division.
(6) Detecting and repairing errors and damage to the genome.
(7) Restructuring the genome when necessary (as part of the normal life cycle or in response to a critical selective challenge).
These additional capabilities involve specific kinds of interactions between DNA and other cellular molecules. The construction of highly precise transcription
complexes in RNA and protein synthesis is one example of such interactions (Ptashne, 1986). Formation of a kinetochore structure at the centromere for attachment of microtubulues to ensure chromosome distribution at mitosis is another example (Volpe et al., 2003).
The idea that repetitive DNA is ‘junk’ without functional significance in the genome is simply not consistent with an extensive and growing literature, only a minor part of which is cited here.
Transposable elements CONTAIN SIGNALS that define the boundaries of each element and help create nucleoprotein structures that allow them to interact with and rearrange target DNA sequences [e.g. terminal inverted repeats (TIRs) of DNA transposons and long terminal repeats (LTRs) in retrotransposons (Craig et al., 2002)]. In addition, transposons and retrotransposons CONTROL SEQUENCE COMPONENTS that control transcription and may participate in DNA replication and chromatin organisation. We know from an extensive literature on insertional mutagenesis in nature and the laboratory that introduction of a transposable element into a particular location CONFERS NEW FUNCTIONAL PROPERTIES on that region of the genome (Shapiro, 1983; Craig et al., 2002; Deininger et al., 2003).
Since gypsy and other mobile elements retain their structures as they migrate through the genome, there is predictability to the signals they will carry with them. Thus, cells have the ability to introduce a PREORGANISED CONSTELLATION OF FUNCTIONAL SIGNALS into any location in the genome.
The notion that LINE elements are MAJOR ORGANISERS of genome functional architecture is supported by close comparative analysis of syntenic regions in the mouse and human genomes.
CONTAIN SIGNALS
CONTROL SEQUENCE COMPONENTS
CONFER NEW FUNCTIONAL PROPERTIES
PREORGANISED CONSTELLATION OF FUNCTIONAL SIGNALS
MAJOR ORGANISERS
Primate specific retrotransposons, SVAs, in the evolution of networks that alter brain function. 2
Our analysis suggests a potential role of SVAs in evolution of human CNS and especially emergence of functional trends relevant to social and parental behaviour. It also supports models which explain in part how brain function can be modulated by both the immune and reproductive systems based on the gene expression patterns and gene pathways potentially altered by SVA insertions.
http://www.icr.org/article/transposable-elements-are-key-genome/
1. http://shapiro.bsd.uchicago.edu/Shapiro&Sternberg.2005.BiolRevs.pdf
2. https://arxiv.org/ftp/arxiv/papers/1602/1602.07642.pdf
Last edited by Admin on Mon Dec 17, 2018 9:52 am; edited 5 times in total