How Genomes ( supposedly ) evolve
Genes with similar functions can be found in a diverse range of living things. But the great revelation of the past 30 years has been the extent to which the actual nucleotide sequences of many genes have been conserved. Homologous genes—that is, genes that are similar in both their nucleotide sequence and function —can often be recognized across vast phylogenetic distances. Unmistakable homologs of many human genes are present in organisms as diverse as nematode worms, fruit flies, yeasts, and even bacteria. In many cases, the resemblance is so close that, for example, the protein-coding portion of a yeast gene can be substituted with its
human homolog. The recognition of sequence similarity has become a major tool for inferring gene and protein function. Although a sequence match does not guarantee similarity in function, it has proved to be an excellent
clue. Thus, it is often possible to predict the function of genes in humans for which no biochemical or genetic information is available simply by comparing their nucleotide sequences with the sequences of genes that have been characterized in other more readily studied organisms. In general, the sequences of individual genes are much more tightly conserved than is overall genome structure. Features of genome organization such as genome size, number of chromosomes, order of genes along chromosomes, abundance and size of introns, and amount of repetitive DNA are found to differ greatly when comparing distant organisms, as does the number of genes that each
organism contains.
Genome Comparisons Reveal Functional DNA Sequences
A first obstacle in interpreting the sequence of the 3.2 billion nucleotide pairs in the human genome is the fact that much of it is probably functionally unimportant. The regions of the genome that code for the amino acid sequences of proteins (the exons) are typically found in short segments (average size about 145 nucleotide pairs), small islands in a sea of DNA whose exact nucleotide sequence is thought to be mostly of little consequence. This arrangement makes it difficult to identify all the exons in a stretch of DNA, and it is often hard too to determine exactly where a gene begins and ends. One very important approach to deciphering our genome is to search for DNA sequences that are closely similar between different species, on the principle that DNA sequences that have a function are much more likely to be conserved than those without a function. In addition to revealing those DNA sequences that encode functionally important exons and RNA molecules, these conserved regions will include regulatory DNA sequences as well as DNA sequences with functions that are not yet known. In contrast, most nonconserved regions will reflect DNA whose sequence is much less likely to be critical for function. The power of this method can be increased by including in such comparisons the genomes of large numbers of species whose genomes have been sequenced, such as rat, chicken, fish, dog, and chimpanzee, as well as mouse and human. Roughly 5% of the human genome consists of “multispecies conserved sequences.” To our great surprise, only about one-third of these sequences code for proteins. Many of the remaining conserved sequences consist of DNA containing clusters of protein-binding sites that are involved in gene regulation, while others produce RNA molecules that are not translated into protein but are important for other known purposes. But, even in the most intensively studied species, the function of the majority of these highly conserve sequences remains unknown. This remarkable discovery has led scientists to conclude that we understand much less about the cell biology of vertebrates than we had thought. Certainly, there are enormous opportunities for new discoveries, and we should expect many more surprises ahead.
Genome Alterations Are Caused by Failures of the Normal Mechanisms for Copying and Maintaining DNA, as well as by Transposable DNA Elements
Evolution depends on accidents and mistakes followed by nonrandom survival. Most of the genetic changes that occur result simply from failures in the normal mechanisms by which genomes are copied or repaired when damaged, although the movement of transposable DNA elements also plays an important part. The mechanisms that maintain DNA sequences are remarkably precise—but they are not perfect. DNA sequences are inherited with such extraordinary fidelity that typically, along a given line of descent, only about one nucleotide pair in a thousand is randomly changed in the germ line every supposed million years. Even so, in a population of 10,000 diploid individuals, every possible nucleotide substitution will have been “tried out” on about 20 occasions in the course of a million years—a short span of time in relation to the supposed evolution of species. Errors in DNA replication, DNA recombination, or DNA repair can lead either to simple local changes in DNA sequence—so-called point mutations such as the substitution of one base pair for another—or to large-scale genome rearrangements such as deletions, duplications, inversions, and translocations of DNA from one chromosome to another. In addition to these failures of the genetic machinery, genomes contain mobile DNA elements that are an important source of genomic change. These transposable DNA elements (transposons) are parasitic DNA sequences that can spread within the genomes they colonize. In the process, they often disrupt the function or alter the regulation of existing genes. On occasion, they have created altogether novel genes through fusions between transposon sequences and segments of existing genes. Over long periods of evolutionary time, DNA transposition events have profoundly affected genomes, so much so that nearly half of the DNA in the human genome consists of recognizable relics of past transposition events .
The Genome Sequences of Two Species Differ in Proportion to the Length of Time Since They Have Separately Evolved
The differences between the genomes of species alive today have supposedly accumulated over more than 3 billion years. Although we lack a direct record of changes over time, scientists can reconstruct the process of genome evolution from detailed comparisons of the genomes of contemporary organisms. The basic organizing framework for comparative genomics is the phylogenetic tree. A simple example is the tree describing the divergence of humans from the great apes
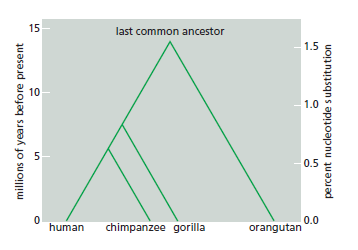
A phylogenetic tree showing the relationship between humans and the great apes based on nucleotide sequence data. As indicated, the sequences of the genomes of all four species are estimated to differ from the sequence of the genome of their last common ancestor by a little over 1.5%. Because changes occur independently on both diverging lineages, pairwise comparisons reveal twice the sequence divergence from the last common ancestor. For example, human–orangutan comparisons typically show sequence
divergences of a little over 3%, while human–chimpanzee comparisons show divergences of approximately 1.2%.
The close similarity between human and chimpanzee genes is mainly due to the short time that has been available for the accumulation of mutations in the two diverging lineages, rather than to functional constraints that have kept the sequences the same. Evidence for this view comes from the observation that the human and chimpanzee genomes are nearly identical even where there is no functional constraint on the nucleotide sequence—such as in the third position of “synonymous” codons (codons specifying the same amino
acid but differing in their third nucleotide). For much less closely related organisms, such as humans and chickens, the sequence conservation found in genes is almost entirely due to purifying selection (that is, selection that eliminates individuals carrying mutations that interfere with important genetic functions), rather than to an inadequate time for mutations to occur.
Phylogenetic Trees Constructed from a Comparison of DNA Sequences Trace the Relationships of All Organisms
Phylogenetic trees based on molecular sequence data can be compared with the fossil record, and we get our best view of evolution by integrating the two approaches. The fossil record remains essential as a source of absolute dates based on radioisotope decay in the rock formations in which fossils are found. Because the fossil record has many gaps, however, precise divergence times between species are difficult to establish, even for species that leave good fossils with distinctive morphology. Phylogenetic trees whose timing has been calibrated according to the fossil record suggest that changes in the sequences of particular genes or proteins tend to occur at a nearly constant rate, although rates that differ from the norm by as much as twofold are observed in particular lineages. This provides us with a molecular clock for evolution—or rather a set of molecular clocks corresponding to different categories of DNA sequence.
Genes with similar functions can be found in a diverse range of living things. But the great revelation of the past 30 years has been the extent to which the actual nucleotide sequences of many genes have been conserved. Homologous genes—that is, genes that are similar in both their nucleotide sequence and function —can often be recognized across vast phylogenetic distances. Unmistakable homologs of many human genes are present in organisms as diverse as nematode worms, fruit flies, yeasts, and even bacteria. In many cases, the resemblance is so close that, for example, the protein-coding portion of a yeast gene can be substituted with its
human homolog. The recognition of sequence similarity has become a major tool for inferring gene and protein function. Although a sequence match does not guarantee similarity in function, it has proved to be an excellent
clue. Thus, it is often possible to predict the function of genes in humans for which no biochemical or genetic information is available simply by comparing their nucleotide sequences with the sequences of genes that have been characterized in other more readily studied organisms. In general, the sequences of individual genes are much more tightly conserved than is overall genome structure. Features of genome organization such as genome size, number of chromosomes, order of genes along chromosomes, abundance and size of introns, and amount of repetitive DNA are found to differ greatly when comparing distant organisms, as does the number of genes that each
organism contains.
Genome Comparisons Reveal Functional DNA Sequences
A first obstacle in interpreting the sequence of the 3.2 billion nucleotide pairs in the human genome is the fact that much of it is probably functionally unimportant. The regions of the genome that code for the amino acid sequences of proteins (the exons) are typically found in short segments (average size about 145 nucleotide pairs), small islands in a sea of DNA whose exact nucleotide sequence is thought to be mostly of little consequence. This arrangement makes it difficult to identify all the exons in a stretch of DNA, and it is often hard too to determine exactly where a gene begins and ends. One very important approach to deciphering our genome is to search for DNA sequences that are closely similar between different species, on the principle that DNA sequences that have a function are much more likely to be conserved than those without a function. In addition to revealing those DNA sequences that encode functionally important exons and RNA molecules, these conserved regions will include regulatory DNA sequences as well as DNA sequences with functions that are not yet known. In contrast, most nonconserved regions will reflect DNA whose sequence is much less likely to be critical for function. The power of this method can be increased by including in such comparisons the genomes of large numbers of species whose genomes have been sequenced, such as rat, chicken, fish, dog, and chimpanzee, as well as mouse and human. Roughly 5% of the human genome consists of “multispecies conserved sequences.” To our great surprise, only about one-third of these sequences code for proteins. Many of the remaining conserved sequences consist of DNA containing clusters of protein-binding sites that are involved in gene regulation, while others produce RNA molecules that are not translated into protein but are important for other known purposes. But, even in the most intensively studied species, the function of the majority of these highly conserve sequences remains unknown. This remarkable discovery has led scientists to conclude that we understand much less about the cell biology of vertebrates than we had thought. Certainly, there are enormous opportunities for new discoveries, and we should expect many more surprises ahead.
Genome Alterations Are Caused by Failures of the Normal Mechanisms for Copying and Maintaining DNA, as well as by Transposable DNA Elements
Evolution depends on accidents and mistakes followed by nonrandom survival. Most of the genetic changes that occur result simply from failures in the normal mechanisms by which genomes are copied or repaired when damaged, although the movement of transposable DNA elements also plays an important part. The mechanisms that maintain DNA sequences are remarkably precise—but they are not perfect. DNA sequences are inherited with such extraordinary fidelity that typically, along a given line of descent, only about one nucleotide pair in a thousand is randomly changed in the germ line every supposed million years. Even so, in a population of 10,000 diploid individuals, every possible nucleotide substitution will have been “tried out” on about 20 occasions in the course of a million years—a short span of time in relation to the supposed evolution of species. Errors in DNA replication, DNA recombination, or DNA repair can lead either to simple local changes in DNA sequence—so-called point mutations such as the substitution of one base pair for another—or to large-scale genome rearrangements such as deletions, duplications, inversions, and translocations of DNA from one chromosome to another. In addition to these failures of the genetic machinery, genomes contain mobile DNA elements that are an important source of genomic change. These transposable DNA elements (transposons) are parasitic DNA sequences that can spread within the genomes they colonize. In the process, they often disrupt the function or alter the regulation of existing genes. On occasion, they have created altogether novel genes through fusions between transposon sequences and segments of existing genes. Over long periods of evolutionary time, DNA transposition events have profoundly affected genomes, so much so that nearly half of the DNA in the human genome consists of recognizable relics of past transposition events .
The Genome Sequences of Two Species Differ in Proportion to the Length of Time Since They Have Separately Evolved
The differences between the genomes of species alive today have supposedly accumulated over more than 3 billion years. Although we lack a direct record of changes over time, scientists can reconstruct the process of genome evolution from detailed comparisons of the genomes of contemporary organisms. The basic organizing framework for comparative genomics is the phylogenetic tree. A simple example is the tree describing the divergence of humans from the great apes

A phylogenetic tree showing the relationship between humans and the great apes based on nucleotide sequence data. As indicated, the sequences of the genomes of all four species are estimated to differ from the sequence of the genome of their last common ancestor by a little over 1.5%. Because changes occur independently on both diverging lineages, pairwise comparisons reveal twice the sequence divergence from the last common ancestor. For example, human–orangutan comparisons typically show sequence
divergences of a little over 3%, while human–chimpanzee comparisons show divergences of approximately 1.2%.
The close similarity between human and chimpanzee genes is mainly due to the short time that has been available for the accumulation of mutations in the two diverging lineages, rather than to functional constraints that have kept the sequences the same. Evidence for this view comes from the observation that the human and chimpanzee genomes are nearly identical even where there is no functional constraint on the nucleotide sequence—such as in the third position of “synonymous” codons (codons specifying the same amino
acid but differing in their third nucleotide). For much less closely related organisms, such as humans and chickens, the sequence conservation found in genes is almost entirely due to purifying selection (that is, selection that eliminates individuals carrying mutations that interfere with important genetic functions), rather than to an inadequate time for mutations to occur.
Phylogenetic Trees Constructed from a Comparison of DNA Sequences Trace the Relationships of All Organisms
Phylogenetic trees based on molecular sequence data can be compared with the fossil record, and we get our best view of evolution by integrating the two approaches. The fossil record remains essential as a source of absolute dates based on radioisotope decay in the rock formations in which fossils are found. Because the fossil record has many gaps, however, precise divergence times between species are difficult to establish, even for species that leave good fossils with distinctive morphology. Phylogenetic trees whose timing has been calibrated according to the fossil record suggest that changes in the sequences of particular genes or proteins tend to occur at a nearly constant rate, although rates that differ from the norm by as much as twofold are observed in particular lineages. This provides us with a molecular clock for evolution—or rather a set of molecular clocks corresponding to different categories of DNA sequence.


