The Mystery of Extreme Non-Coding Conservation 2
The argument of the highly similar DNA sequences
1. If functionally unconstrained yet highly similar DNA sequences were found in different species, then evolution would be false.
2. In fact, the DNA sequences are extremely similar and even identical in different species.
3. There is currently “no known mechanism or function that would account for this level of conservation at the observed evolutionary distances.”
4. Since some of these sequences are found across a wide range of different species, the sequences, and whatever selective forces preserved them, must have been present very early in history.
5. On the other hand many of these sequences point to evolution’s nemesis, lineage-specific biology.
6. Highly similar DNA sequences in different species are a proof of the same intelligent designer using a similar genetic pattern to design different species. All men call him God.
7. God exists.
Evolution is unique in that while it is well known amongst evolutionists to be a fact, its predictions often turn out false. Consider this new paper from the Royal Society on “The mystery of extreme non-coding conservation” that has been found across many genomes. Years ago an evolution professor told me, in defending the claim that evolution is falsifiable, that if functionally unconstrained yet highly similar DNA sequences were found in different species, then evolution would be false. A few years later that is exactly what was discovered. In fact, the DNA sequences were extremely similar and even identical in different species, and when they were altogether removed from mice it made no detectable difference. Hundreds of tests showed no significant difference between mice with and without long stretches of these DNA sequences. Did the professor agree that evolution was false? Not at all. For the fact of evolution goes far deeper than scientific findings and failed predictions. Nonetheless, ten years later, the mystery of extreme DNA conservation remains.
As the paper explains, there is currently “no known mechanism or function that would account for this level of conservation at the observed evolutionary distances.” This failure forces us to draw upon the typical explanatory mechanisms. The evolution of these extremely conserved sequences must have been abrupt and rapid, occurring in “short bursts.”
And since some of these sequences are found across a wide range of different species, the sequences, and whatever selective forces preserved them, must have been present very early in evolutionary history. On the other hand many of these sequences point to evolution’s nemesis, lineage-specific biology.
Some of these sequences are extremely conserved within lineages, but not across lineages. This forces us to conclude that the ancestral sequence first somehow arose in the common ancestor, later evolved independently in the different lineages which arose, became completely different in those different lineages, and then finally each of these different sequences, in the respective lineages, somehow became essentially unchangeable.
As is typical of the evolution genre, all of this is expressed in teleological terms. Here is a paragraph from the paper that is loaded with evolution’s Aristotelian tendencies:
Lowe et al. proposed that, within vertebrates, there have been three distinct periods of CNE [conserved non-coding element] recruitment around specific groups of genes. They suggest that this pattern is the result of regulatoryinnovations, which led to important phenotypic changes during vertebrate evolution. Prior to the divergence of mammals from reptiles and birds, it appears that CNEs were preferentially recruited near TFs and their developmental targets. This was followed by a gradual decline in recruitmentnear these genes, accompanied by [a recruitment] increase near proteins involved in extracellular signalling, and then [a recruitment] increase in placental mammals near genes responsible for post-translational modification and intracellular signalling. An analysis of CNE gain in the primate and rodent lineage has found that CNEs are either recruited near genes which have not previously been associated with CNEs, or are addednear genes which are already flanked by CNEs. The interpretation was that the first set of genes is enriched in functions pertaining to nervous system development, whereas the latter contains genes involved in transcriptional regulation and anatomical development.
This example of teleological language also illustrates how the commitment to a theory can lead to a loss of parsimony. That is, in order to accommodate new and contradictory findings, additional explanations must be added to the theory. It becomes more complicated and less parsimonious. Here is how the paper summarizes these findings of extreme sequence conservation:
… despite 10 years of research, there has been virtually no progress towards answering the question of the origin of these patterns of extreme conservation. A number of hypotheses have been proposed, but most rely on modes of DNA : protein interactions that have never been observed and seem dubious at best. As a consequence, not only do we still lack a plausible mechanism for the conservation of CNEs—we lack even plausible speculations.
Reasonable speculation and even solutions to extreme sequence conservation may come in the future. But today’s science once again highlights the unique status of evolution.
The Developmental Genetic Toolkit and the Molecular Homology–Analogy Paradox 1
It came as a big surprise to workers in the fields of evolutionary and developmental biology to learn that the Drosophila eyeless (ey) gene, mutations of which cause loss or reduction in size of eyes, has extensive DNA sequence similarity to the mouse and human Pax-6 genes, which when mutated reduce the size of the mammalian eye. Before this unexpected finding, insect and vertebrate eyes were believed to be analogues—independently evolved structures playing similar or corresponding functions—not homologues—structures whose similarity is based on a common ancestral prototype. Insects have compound eyes with numerous microscopic units (ommatidia), each with its own tiny lens and set of photosensitive cells, whereas the eyes of mammals and other vertebrates are organized like cameras, with a single macroscopic lens projecting onto an extended, planar retina. Moreover, arthropods and chordates, the larger taxonomic groups to which insects and vertebrates belong, began following separate trajectories . Drosophila eyeless and vertebrate Pax-6 are both what are termed “master control genes,” in that they specify transcription factors that stand at the apex of particular developmental pathways. Other such factors initiate pathways leading to muscle and cartilage in vertebrates and glial cells in Drosophila. Twin of eyeless is a Drosophila gene that acts upstream of ey and thus fits the description of master control gene for eye development even better than the latter. Significantly, it is even more closely related to vertebrate Pax-6 than is ey. It might be expected that eyes across wide taxonomic distances, even those that may have evolved independently, would employ common “nuts and bolts,” e.g., photosensitive pigments and electrically excitable channels. But given the vastly different developmental processes leading to the compound and camera eyes, it was extremely puzzling to find that the same transcription factor would act as a master control molecule in such morphologically distinct organs in such evolutionarily distant taxa. As Walter Gehring, who discovered the correspondence between ey and Pax-6, noted: “. . . there is no functional necessity to use a particular transcription factor like Pax 6 for particular function e.g. eye morphogenesis, since a transcription factor can regulate any gene, if this gene is endowed with the appropriate regulatory elements in its enhancer or promoter” . The discovery of the similarity between arthropod and chordate master control genes for eye development, as well as related genes in other distant taxonomic groups in which eyes are present (e.g., cephalopod molluscs;, has led to suggestions that these various eyes are indeed homologous. Muller, in contrast, drawing on a concept of homology ¨that focuses on organizational units (e.g., constructional elements or phenotypic characters), concluded that despite the use of common gene regulatory pathways arthropod, mollusc, and chordate eyes are decidedly not anatomically homologous. In general, new molecular knowledge has reinforced the notion of ambiguity in assignments of homology and analogy. In what follows I will refer to the set of problems raised by the use of homologous genes in the generation of what have classically been considered to be analogous structures in different phyla as the “molecular homology–analogy (MHA) paradox”.
Another example of the MHA paradox involves segmentation, the formation of sequentially repeated tissue modules along the body axis. This process occurs in arthropods, annelids, and vertebrates, which are not thought to have a last common ancestor that was segmented. William Bateson considered that segmentation arose independently at least a half dozen times and since then it has widely been considered a classic example of “homoplasy,” ( convergent evolution ) similarity of characters for reasons other than common descent , that is, parallel or convergent evolution. Nonetheless, certain genes specifying homologous transcription factors of the hairy/enhancer of split (Hes) class in Drosophila, in the annelid leech Helobdella robusta, and in vertebrates are causally involved in segmentation by virtue of being expressed in a periodic fashion: in the case of Drosophila, spatially periodic, and in the leech and vertebrate, temporally periodic. Since any periodically expressed transcription factor could potentially do the job—mediating the formation of boundaries between blocks of tissue —Gehring’s observation concerning ey/Pax-6 quoted above holds with equal force
Other examples abound. The homologous genes extradenticle and Pbx1 are involved in specifying proximal regions of appendages in Drosophila and vertebrates, respectively ,whereas the homologues Distal-less in insects and Dlx in vertebrates are involved in distal specification. This is despite the fact that insect and vertebrate appendages have little in common anatomically other than being produced, in part, by outgrowth of the body surface. Formation of the pharyngeal muscles of the nematode Caenorhabditis elegans requires the expression of the homeobox gene ceh-22, which is homologous to genes of the NK-2 family involved in heart development in insects and vertebrates . In addition to these organ-, segmental-, and proximaland distal-specific master genes (along with their associated pathways) being propagated among disparate forms during the wholesale generation and reconfiguration of body plans of metazoan diversification, a small number of fundamental receptor-ligand-based intercellular signaling systems that apparently arose even earlier, such as the Notch-Delta ligand receptor pair and the Wntcatenin pathway, also continued to be ubiquitously involved in local differentiation and tissue regionalization processes during the origination of metazoan phyla. The molecules initiating these pathways (Notch, Wnt), together with the ones mentioned above and a few others (e.g., Hedgehog, T-box, and Hox proteins), have been referred to as the conserved metazoan developmental genetic toolkit.
Although the discovery of the pan-metazoan toolkit was initially met with astonishment , it was subsequently assimilated into the standard neo-Darwinian framework by being pointed to as confirmation of the common molecular roots of the tree of life and repeated cooptation of evolution ary related molecules for similar purposes . These observations do not resolve the MHA paradox, however. The molecular commonality of biological
systems has long been acknowledged: the use of DNA and/or RNA as genetic molecules, the genetic code, and the use of proteins and RNA molecules as chemical catalysts are common to all cellular life. Histones, actin, and tubulin, proteins with irreplaceable functions in individual eukaryotic cells, are homologous in animals, plants, and fungi. While there would have been a high cost in changing any of these essential constituents of eukaryotic life once multicellularity arose, there should have been little constraint on the proteins used to initiate and coordinate analogous developmental pathways, as noted by Gehring above. Comparison of animals and plants, for example, has led to the conclusion that their basic mechanisms of pattern formation and of cell–cell communication in development were independently derived.
The major reason that the use of homologous molecules to serve anatomically and functionally similar purposes seems paradoxical is that evolutionary theory has traditionally held that very long periods of time were needed for natural selection to generate extreme differences in morphological organization like those seen in animal body plans. In particular, the neo-Darwinian mechanism for such large-scale change was the accumulation, along multiple evolutionary trajectories, of incremental gene-related (i.e., microevolutionary) departures from a common ancestral morphology. This view was shaken by the recognition over the last quarter century that essentially all modern metazoan body plans burst into the biosphere over a relatively short period of time, geologically speaking, perhaps 20–30 million years, during the late pre-Cambrian and early Cambrian periods about 530–570 million years ago. The so-called Cambrian explosion has been likened to the cosmic big bang in its concentration of enormous change over an extraordinarily brief time span.
Various interpretations, generally conforming to the neoDarwinian picture, have been put forward for the unexpected rate of morphological evolution at the dawn of the metazoa. When the MHA paradox is considered in relation to the suggested scenarios for the Cambrian explosion, however, certain inconsistencies emerge that suggest additional causal agency is required. One scenario is that the selective environment was so intense
at the origin of the metazoa that microevolution was accelerated substantially over the usual rate. Presumably this rapid evolution mainly affected promoters, enhancers, and other regulatory sequences of DNA, since the sequences of organ-related transcription factors like those mentioned above were left comparatively unchanged over the period of feverish natural selection. But if evolution was intense enough to generate, by small, incremental changes, arthropods, annelids, echinoderms, molluscs, nematodes, and chordates from a common, presumably simpler, ancestor, why were the same transcription factors repeatedly independently recruited to make similar structures in these radically different forms? Moreover, the assumption that alteration of regulatory sequences is sufficient to turn a structurally simple ancestral form (although one with latent appendages, segments, eyes, heart) into forms as varied as flies, sea urchins, nematodes, and humans is widely accepted but difficult to prove. It usually goes unquestioned, since the motive force in the standard model of phenotypic
evolution is DNA change, and in the cases under consideration coding sequences are essentially fixed. In fact, this supposition is not even testable at present since so little is known about the grammar of developmental gene regulation.
An alternative proposal for resolution of the MHA paradox is that extensive evolution occurred prior to the Cambrian explosion, leading to organismal forms similar to the early larvae of indirect developing sea urchins. These microscopic organisms (problematically, not expected to leave much of a trace in the fossil record) are hypothesized to have developed using local cell–cell interactions based on the Notch and Wnt pathways, like modern sea urchin larvae. According to this view, more complex metazoa arose later, from “set-aside” cells that originally emerged in certain of these larva-like forms. This model purports to solve the MHA paradox by suggesting that the toolkit genes were originally components of early-evolved cell differentiation pathways. Once set-aside cells arose, morphogenetic mechanisms recruited the same pathways into functionally analogous organs . While this is plausible for the NK-2 class genes in heart muscle development, it is less so for the Pax-6 class genes and eye development, where more than cell differentiation is involved. For the Hes class genes involved in segmentation, or the Pbx1 and Dlx class genes involved in proximal and distal appendage specification, where the common regulatory factors are not linked to common cell types, the differentiation first model lacks plausibility. It is also not clear why, on this hypothesis, set-aside cells, and the organisms proposed to be evolutionarily based on them, would not have been freed up to invent new local cell-cell signaling systems analogous to the Notch and Wnt pathways. While sea urchins, invertebrate deuterostomes, have extensive genetic affinity with chordates, they are actually outliers in terms of the way they employ the developmental gene networks mentioned above . Moreover, not having eyes, segments or hearts, echinoderms would seem to have only limited relevancy to the MHA paradox as described here.
So the question remains: What kind of evolutionary process could rapidly generate a collection of morphologically distinct metazoan body plans from a common ancestor of annelids, molluscs, nematodes, arthropods, and chordates, while maintaining largely unchanged a core developmental toolkit? I would like to propose that a mechanism based on the plasticity of development , rather than on incremental deviations from an ancestral phenotype, has the potential to resolve this question. We have suggested previously that such plasticity may have been much greater in early multicellular forms than it is in modern forms, by virtue of the fact that the high degree of “overdetermination” characteristic of modern developmental systems would not have been present . Under these “pre- ¨ canalized” conditions, the (relatively unconstrained) physical propensity of viscoelastic, chemically and mechanically excitable, multicellular aggregates to form hollow, tubular, multilayered, and segmented structures would have resulted in a broad but delimited range of bauplans . Here comes the mambo jambo pseudo scientific part in order to keep the naturalistic view and press the evidence in order that the ToE can be kept alive.
In this hypothesized “pre-Mendelian, pre-Darwinian” world of protean organisms, many interconvertible morphological phenotypes would likely have been associated with one or a few basic genotypes. While the organisms’ inherent material properties, and external conditions, would have had more influence over morphogenesis than at present, random mutation, followed by canalizing or stabilizing evolution that reinforced one or another of the multifarious possible morphologies, would have set metazoan life on the path to taxonomic modernity.
While necessarily speculative, this mechanism has the benefit of being rapid, consistent with known molecular genetic and physical processes, and capable of generating highly disparate morphologies. It has no problem, for example, in accounting for the problematic “inversion” of the dorsoventral axis that distinguishes protostomes from deuterostomes . While it is, strictly speaking, neither Mendelian nor Darwinian, it is not difficult to visualize how modern-day metazoa that are both Mendelian and Darwinian could have evolved from the hypothesized primitive organisms. Significantly, by invoking physical and other epigenetic determinants, rather than genetic change, to account for the burst of metazoan body plans in the transition from the pre-Cambrian to the Cambrian period, this conjecture affords an understanding of how such morphological diversification could have taken place without altering the primordial molecular toolkit, and thus a possible resolution of the MHA paradox.
THE HIERARCHICAL ORGANIZATION OF GENETIC AND EPIGENETIC INFORMATION
Stephen C Meyer , Darwin's doubt pg.268:
There is another remarkable aspect of the hierarchical organization of information in animal forms. Many of the same genes and proteins play very different roles, depending upon the larger organismal and informational context in which they find themselves in different animal groups. For example, the same gene (Pax-6 or its homolog, called eyeless), helps to regulate the development of the eyes of fruit flies (arthropods) and those of squid and mice (cephalopods and vertebrates, respectively). Yet arthropod eyes exemplify a completely different structure from vertebrate or cephalopod eyes. The fruit fly possesses a compound eye with hundreds of separate lenses (ommatidia), whereas both mice and squid employ a camera-type eye with a single lens and retinal surface. In addition, although the eyes of squid and mice resemble each other optically (single lens, large internal chamber, single retinal surface), they focus differently. They undergo completely different patterns of development and utilize different internal structures and nerve connections to the visual centers of the brain. Yet the Pax-6 gene and its homologs play a key role in regulating the construction of all three of these different adult sensory structures. Moreover, evolutionary and developmental biologists have found that this pattern of "same genes, different anatomy" recurs throughout the bilaterian phyla, for features as fundamental as appendages, segmentation, the gut, heart, and sense organs
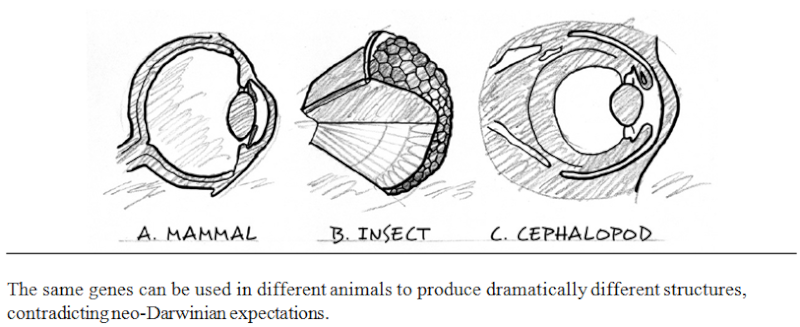
This pattern contradicts the expectations of textbook evolutionary theory. Neo-Darwinism predicts that disparate adult structures should be produced by different genes. This prediction follows directly from the neo-Darwinian assumption that all evolutionary (including anatomical) transformations begin with mutations in DNA sequences—mutations that are fixed in populations by natural selection, genetic drift, or other evolutionary processes. The arrow of causality flows one way from genes (DNA) to development to adult anatomy. Thus, if biologists observe different animal forms, it follows that they should expect that different genes will specify those forms during animal development. Given the profound differences between the fruit-fly compound eye and the vertebrate camera eye, neo-Darwinian theory would not predict that the "same" genes would be involved in building different eyes in arthropods and chordates.
1) https://www.nymc.edu/sanewman/PDFs/Developmental-genetic%20toolkit.pdf
2) http://darwins-god.blogspot.com.br/2013/11/the-mystery-of-extreme-non-coding.html
The argument of the highly similar DNA sequences
1. If functionally unconstrained yet highly similar DNA sequences were found in different species, then evolution would be false.
2. In fact, the DNA sequences are extremely similar and even identical in different species.
3. There is currently “no known mechanism or function that would account for this level of conservation at the observed evolutionary distances.”
4. Since some of these sequences are found across a wide range of different species, the sequences, and whatever selective forces preserved them, must have been present very early in history.
5. On the other hand many of these sequences point to evolution’s nemesis, lineage-specific biology.
6. Highly similar DNA sequences in different species are a proof of the same intelligent designer using a similar genetic pattern to design different species. All men call him God.
7. God exists.
Evolution is unique in that while it is well known amongst evolutionists to be a fact, its predictions often turn out false. Consider this new paper from the Royal Society on “The mystery of extreme non-coding conservation” that has been found across many genomes. Years ago an evolution professor told me, in defending the claim that evolution is falsifiable, that if functionally unconstrained yet highly similar DNA sequences were found in different species, then evolution would be false. A few years later that is exactly what was discovered. In fact, the DNA sequences were extremely similar and even identical in different species, and when they were altogether removed from mice it made no detectable difference. Hundreds of tests showed no significant difference between mice with and without long stretches of these DNA sequences. Did the professor agree that evolution was false? Not at all. For the fact of evolution goes far deeper than scientific findings and failed predictions. Nonetheless, ten years later, the mystery of extreme DNA conservation remains.
As the paper explains, there is currently “no known mechanism or function that would account for this level of conservation at the observed evolutionary distances.” This failure forces us to draw upon the typical explanatory mechanisms. The evolution of these extremely conserved sequences must have been abrupt and rapid, occurring in “short bursts.”
And since some of these sequences are found across a wide range of different species, the sequences, and whatever selective forces preserved them, must have been present very early in evolutionary history. On the other hand many of these sequences point to evolution’s nemesis, lineage-specific biology.
Some of these sequences are extremely conserved within lineages, but not across lineages. This forces us to conclude that the ancestral sequence first somehow arose in the common ancestor, later evolved independently in the different lineages which arose, became completely different in those different lineages, and then finally each of these different sequences, in the respective lineages, somehow became essentially unchangeable.
As is typical of the evolution genre, all of this is expressed in teleological terms. Here is a paragraph from the paper that is loaded with evolution’s Aristotelian tendencies:
Lowe et al. proposed that, within vertebrates, there have been three distinct periods of CNE [conserved non-coding element] recruitment around specific groups of genes. They suggest that this pattern is the result of regulatoryinnovations, which led to important phenotypic changes during vertebrate evolution. Prior to the divergence of mammals from reptiles and birds, it appears that CNEs were preferentially recruited near TFs and their developmental targets. This was followed by a gradual decline in recruitmentnear these genes, accompanied by [a recruitment] increase near proteins involved in extracellular signalling, and then [a recruitment] increase in placental mammals near genes responsible for post-translational modification and intracellular signalling. An analysis of CNE gain in the primate and rodent lineage has found that CNEs are either recruited near genes which have not previously been associated with CNEs, or are addednear genes which are already flanked by CNEs. The interpretation was that the first set of genes is enriched in functions pertaining to nervous system development, whereas the latter contains genes involved in transcriptional regulation and anatomical development.
This example of teleological language also illustrates how the commitment to a theory can lead to a loss of parsimony. That is, in order to accommodate new and contradictory findings, additional explanations must be added to the theory. It becomes more complicated and less parsimonious. Here is how the paper summarizes these findings of extreme sequence conservation:
… despite 10 years of research, there has been virtually no progress towards answering the question of the origin of these patterns of extreme conservation. A number of hypotheses have been proposed, but most rely on modes of DNA : protein interactions that have never been observed and seem dubious at best. As a consequence, not only do we still lack a plausible mechanism for the conservation of CNEs—we lack even plausible speculations.
Reasonable speculation and even solutions to extreme sequence conservation may come in the future. But today’s science once again highlights the unique status of evolution.
The Developmental Genetic Toolkit and the Molecular Homology–Analogy Paradox 1
It came as a big surprise to workers in the fields of evolutionary and developmental biology to learn that the Drosophila eyeless (ey) gene, mutations of which cause loss or reduction in size of eyes, has extensive DNA sequence similarity to the mouse and human Pax-6 genes, which when mutated reduce the size of the mammalian eye. Before this unexpected finding, insect and vertebrate eyes were believed to be analogues—independently evolved structures playing similar or corresponding functions—not homologues—structures whose similarity is based on a common ancestral prototype. Insects have compound eyes with numerous microscopic units (ommatidia), each with its own tiny lens and set of photosensitive cells, whereas the eyes of mammals and other vertebrates are organized like cameras, with a single macroscopic lens projecting onto an extended, planar retina. Moreover, arthropods and chordates, the larger taxonomic groups to which insects and vertebrates belong, began following separate trajectories . Drosophila eyeless and vertebrate Pax-6 are both what are termed “master control genes,” in that they specify transcription factors that stand at the apex of particular developmental pathways. Other such factors initiate pathways leading to muscle and cartilage in vertebrates and glial cells in Drosophila. Twin of eyeless is a Drosophila gene that acts upstream of ey and thus fits the description of master control gene for eye development even better than the latter. Significantly, it is even more closely related to vertebrate Pax-6 than is ey. It might be expected that eyes across wide taxonomic distances, even those that may have evolved independently, would employ common “nuts and bolts,” e.g., photosensitive pigments and electrically excitable channels. But given the vastly different developmental processes leading to the compound and camera eyes, it was extremely puzzling to find that the same transcription factor would act as a master control molecule in such morphologically distinct organs in such evolutionarily distant taxa. As Walter Gehring, who discovered the correspondence between ey and Pax-6, noted: “. . . there is no functional necessity to use a particular transcription factor like Pax 6 for particular function e.g. eye morphogenesis, since a transcription factor can regulate any gene, if this gene is endowed with the appropriate regulatory elements in its enhancer or promoter” . The discovery of the similarity between arthropod and chordate master control genes for eye development, as well as related genes in other distant taxonomic groups in which eyes are present (e.g., cephalopod molluscs;, has led to suggestions that these various eyes are indeed homologous. Muller, in contrast, drawing on a concept of homology ¨that focuses on organizational units (e.g., constructional elements or phenotypic characters), concluded that despite the use of common gene regulatory pathways arthropod, mollusc, and chordate eyes are decidedly not anatomically homologous. In general, new molecular knowledge has reinforced the notion of ambiguity in assignments of homology and analogy. In what follows I will refer to the set of problems raised by the use of homologous genes in the generation of what have classically been considered to be analogous structures in different phyla as the “molecular homology–analogy (MHA) paradox”.
Another example of the MHA paradox involves segmentation, the formation of sequentially repeated tissue modules along the body axis. This process occurs in arthropods, annelids, and vertebrates, which are not thought to have a last common ancestor that was segmented. William Bateson considered that segmentation arose independently at least a half dozen times and since then it has widely been considered a classic example of “homoplasy,” ( convergent evolution ) similarity of characters for reasons other than common descent , that is, parallel or convergent evolution. Nonetheless, certain genes specifying homologous transcription factors of the hairy/enhancer of split (Hes) class in Drosophila, in the annelid leech Helobdella robusta, and in vertebrates are causally involved in segmentation by virtue of being expressed in a periodic fashion: in the case of Drosophila, spatially periodic, and in the leech and vertebrate, temporally periodic. Since any periodically expressed transcription factor could potentially do the job—mediating the formation of boundaries between blocks of tissue —Gehring’s observation concerning ey/Pax-6 quoted above holds with equal force
Other examples abound. The homologous genes extradenticle and Pbx1 are involved in specifying proximal regions of appendages in Drosophila and vertebrates, respectively ,whereas the homologues Distal-less in insects and Dlx in vertebrates are involved in distal specification. This is despite the fact that insect and vertebrate appendages have little in common anatomically other than being produced, in part, by outgrowth of the body surface. Formation of the pharyngeal muscles of the nematode Caenorhabditis elegans requires the expression of the homeobox gene ceh-22, which is homologous to genes of the NK-2 family involved in heart development in insects and vertebrates . In addition to these organ-, segmental-, and proximaland distal-specific master genes (along with their associated pathways) being propagated among disparate forms during the wholesale generation and reconfiguration of body plans of metazoan diversification, a small number of fundamental receptor-ligand-based intercellular signaling systems that apparently arose even earlier, such as the Notch-Delta ligand receptor pair and the Wntcatenin pathway, also continued to be ubiquitously involved in local differentiation and tissue regionalization processes during the origination of metazoan phyla. The molecules initiating these pathways (Notch, Wnt), together with the ones mentioned above and a few others (e.g., Hedgehog, T-box, and Hox proteins), have been referred to as the conserved metazoan developmental genetic toolkit.
Although the discovery of the pan-metazoan toolkit was initially met with astonishment , it was subsequently assimilated into the standard neo-Darwinian framework by being pointed to as confirmation of the common molecular roots of the tree of life and repeated cooptation of evolution ary related molecules for similar purposes . These observations do not resolve the MHA paradox, however. The molecular commonality of biological
systems has long been acknowledged: the use of DNA and/or RNA as genetic molecules, the genetic code, and the use of proteins and RNA molecules as chemical catalysts are common to all cellular life. Histones, actin, and tubulin, proteins with irreplaceable functions in individual eukaryotic cells, are homologous in animals, plants, and fungi. While there would have been a high cost in changing any of these essential constituents of eukaryotic life once multicellularity arose, there should have been little constraint on the proteins used to initiate and coordinate analogous developmental pathways, as noted by Gehring above. Comparison of animals and plants, for example, has led to the conclusion that their basic mechanisms of pattern formation and of cell–cell communication in development were independently derived.
The major reason that the use of homologous molecules to serve anatomically and functionally similar purposes seems paradoxical is that evolutionary theory has traditionally held that very long periods of time were needed for natural selection to generate extreme differences in morphological organization like those seen in animal body plans. In particular, the neo-Darwinian mechanism for such large-scale change was the accumulation, along multiple evolutionary trajectories, of incremental gene-related (i.e., microevolutionary) departures from a common ancestral morphology. This view was shaken by the recognition over the last quarter century that essentially all modern metazoan body plans burst into the biosphere over a relatively short period of time, geologically speaking, perhaps 20–30 million years, during the late pre-Cambrian and early Cambrian periods about 530–570 million years ago. The so-called Cambrian explosion has been likened to the cosmic big bang in its concentration of enormous change over an extraordinarily brief time span.
Various interpretations, generally conforming to the neoDarwinian picture, have been put forward for the unexpected rate of morphological evolution at the dawn of the metazoa. When the MHA paradox is considered in relation to the suggested scenarios for the Cambrian explosion, however, certain inconsistencies emerge that suggest additional causal agency is required. One scenario is that the selective environment was so intense
at the origin of the metazoa that microevolution was accelerated substantially over the usual rate. Presumably this rapid evolution mainly affected promoters, enhancers, and other regulatory sequences of DNA, since the sequences of organ-related transcription factors like those mentioned above were left comparatively unchanged over the period of feverish natural selection. But if evolution was intense enough to generate, by small, incremental changes, arthropods, annelids, echinoderms, molluscs, nematodes, and chordates from a common, presumably simpler, ancestor, why were the same transcription factors repeatedly independently recruited to make similar structures in these radically different forms? Moreover, the assumption that alteration of regulatory sequences is sufficient to turn a structurally simple ancestral form (although one with latent appendages, segments, eyes, heart) into forms as varied as flies, sea urchins, nematodes, and humans is widely accepted but difficult to prove. It usually goes unquestioned, since the motive force in the standard model of phenotypic
evolution is DNA change, and in the cases under consideration coding sequences are essentially fixed. In fact, this supposition is not even testable at present since so little is known about the grammar of developmental gene regulation.
An alternative proposal for resolution of the MHA paradox is that extensive evolution occurred prior to the Cambrian explosion, leading to organismal forms similar to the early larvae of indirect developing sea urchins. These microscopic organisms (problematically, not expected to leave much of a trace in the fossil record) are hypothesized to have developed using local cell–cell interactions based on the Notch and Wnt pathways, like modern sea urchin larvae. According to this view, more complex metazoa arose later, from “set-aside” cells that originally emerged in certain of these larva-like forms. This model purports to solve the MHA paradox by suggesting that the toolkit genes were originally components of early-evolved cell differentiation pathways. Once set-aside cells arose, morphogenetic mechanisms recruited the same pathways into functionally analogous organs . While this is plausible for the NK-2 class genes in heart muscle development, it is less so for the Pax-6 class genes and eye development, where more than cell differentiation is involved. For the Hes class genes involved in segmentation, or the Pbx1 and Dlx class genes involved in proximal and distal appendage specification, where the common regulatory factors are not linked to common cell types, the differentiation first model lacks plausibility. It is also not clear why, on this hypothesis, set-aside cells, and the organisms proposed to be evolutionarily based on them, would not have been freed up to invent new local cell-cell signaling systems analogous to the Notch and Wnt pathways. While sea urchins, invertebrate deuterostomes, have extensive genetic affinity with chordates, they are actually outliers in terms of the way they employ the developmental gene networks mentioned above . Moreover, not having eyes, segments or hearts, echinoderms would seem to have only limited relevancy to the MHA paradox as described here.
So the question remains: What kind of evolutionary process could rapidly generate a collection of morphologically distinct metazoan body plans from a common ancestor of annelids, molluscs, nematodes, arthropods, and chordates, while maintaining largely unchanged a core developmental toolkit? I would like to propose that a mechanism based on the plasticity of development , rather than on incremental deviations from an ancestral phenotype, has the potential to resolve this question. We have suggested previously that such plasticity may have been much greater in early multicellular forms than it is in modern forms, by virtue of the fact that the high degree of “overdetermination” characteristic of modern developmental systems would not have been present . Under these “pre- ¨ canalized” conditions, the (relatively unconstrained) physical propensity of viscoelastic, chemically and mechanically excitable, multicellular aggregates to form hollow, tubular, multilayered, and segmented structures would have resulted in a broad but delimited range of bauplans . Here comes the mambo jambo pseudo scientific part in order to keep the naturalistic view and press the evidence in order that the ToE can be kept alive.
In this hypothesized “pre-Mendelian, pre-Darwinian” world of protean organisms, many interconvertible morphological phenotypes would likely have been associated with one or a few basic genotypes. While the organisms’ inherent material properties, and external conditions, would have had more influence over morphogenesis than at present, random mutation, followed by canalizing or stabilizing evolution that reinforced one or another of the multifarious possible morphologies, would have set metazoan life on the path to taxonomic modernity.
While necessarily speculative, this mechanism has the benefit of being rapid, consistent with known molecular genetic and physical processes, and capable of generating highly disparate morphologies. It has no problem, for example, in accounting for the problematic “inversion” of the dorsoventral axis that distinguishes protostomes from deuterostomes . While it is, strictly speaking, neither Mendelian nor Darwinian, it is not difficult to visualize how modern-day metazoa that are both Mendelian and Darwinian could have evolved from the hypothesized primitive organisms. Significantly, by invoking physical and other epigenetic determinants, rather than genetic change, to account for the burst of metazoan body plans in the transition from the pre-Cambrian to the Cambrian period, this conjecture affords an understanding of how such morphological diversification could have taken place without altering the primordial molecular toolkit, and thus a possible resolution of the MHA paradox.
THE HIERARCHICAL ORGANIZATION OF GENETIC AND EPIGENETIC INFORMATION
Stephen C Meyer , Darwin's doubt pg.268:
There is another remarkable aspect of the hierarchical organization of information in animal forms. Many of the same genes and proteins play very different roles, depending upon the larger organismal and informational context in which they find themselves in different animal groups. For example, the same gene (Pax-6 or its homolog, called eyeless), helps to regulate the development of the eyes of fruit flies (arthropods) and those of squid and mice (cephalopods and vertebrates, respectively). Yet arthropod eyes exemplify a completely different structure from vertebrate or cephalopod eyes. The fruit fly possesses a compound eye with hundreds of separate lenses (ommatidia), whereas both mice and squid employ a camera-type eye with a single lens and retinal surface. In addition, although the eyes of squid and mice resemble each other optically (single lens, large internal chamber, single retinal surface), they focus differently. They undergo completely different patterns of development and utilize different internal structures and nerve connections to the visual centers of the brain. Yet the Pax-6 gene and its homologs play a key role in regulating the construction of all three of these different adult sensory structures. Moreover, evolutionary and developmental biologists have found that this pattern of "same genes, different anatomy" recurs throughout the bilaterian phyla, for features as fundamental as appendages, segmentation, the gut, heart, and sense organs

This pattern contradicts the expectations of textbook evolutionary theory. Neo-Darwinism predicts that disparate adult structures should be produced by different genes. This prediction follows directly from the neo-Darwinian assumption that all evolutionary (including anatomical) transformations begin with mutations in DNA sequences—mutations that are fixed in populations by natural selection, genetic drift, or other evolutionary processes. The arrow of causality flows one way from genes (DNA) to development to adult anatomy. Thus, if biologists observe different animal forms, it follows that they should expect that different genes will specify those forms during animal development. Given the profound differences between the fruit-fly compound eye and the vertebrate camera eye, neo-Darwinian theory would not predict that the "same" genes would be involved in building different eyes in arthropods and chordates.
1) https://www.nymc.edu/sanewman/PDFs/Developmental-genetic%20toolkit.pdf
2) http://darwins-god.blogspot.com.br/2013/11/the-mystery-of-extreme-non-coding.html
Last edited by Admin on Thu Dec 03, 2015 10:08 pm; edited 6 times in total

