Kinetochores
Kinetochores attach sister chromatids to the Spindle
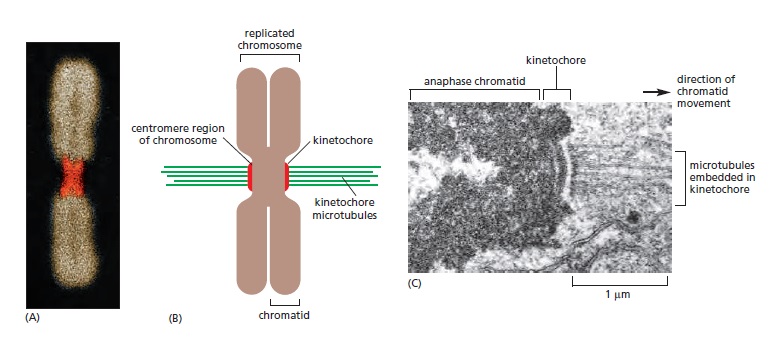
Following the assembly of a bipolar microtubule array, the second major step in spindle formation is the attachment of the array to the sister-chromatid pairs. Spindle microtubules become attached to each chromatid at its kinetochore, a giant, multilayered protein structure that is built at the centromeric region of the chromatid (see picture below)
The kinetochore. see below (A) A fluorescence micrograph of a metaphase chromosome stained with a DNA-binding fluorescent dye and with human autoantibodies that react with specific kinetochore proteins. The two kinetochores, one associated with each sister chromatid, are stained red. (B) A drawing of a metaphase chromosome showing its two sister chromatids attached to the plus ends of kinetochore microtubules. Each kinetochore forms a plaque on the surface of the centromere. (C) Electron micrograph of an anaphase chromatid with microtubules attached to its kinetochore. While most kinetochores have a trilaminar structure, the one shown here (from a green alga) has an unusually complex structure with additional layers.
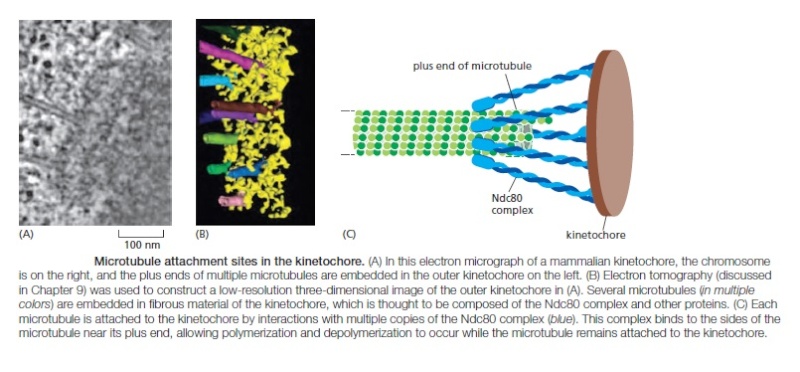
In metaphase, the plus ends of kinetochore microtubules are embedded head-on in specialized microtubuleattachment sites within the outer region of the kinetochore, furthest from the DNA. The kinetochore of an animal cell can bind 10–40 microtubules, whereas a budding yeast kinetochore can bind only one. Attachment of each microtubule depends on multiple copies of a rod-shaped protein complex called the Ndc80 complex, which is anchored in the kinetochore at one end and interacts with the sides of the microtubule at the other, thereby linking the microtubule to the kinetochore while still allowing the addition or removal of tubulin subunits at this end.
Regulation of plus-end polymerization and depolymerization at the kinetochore is critical for the control of chromosome movement on the spindle. Kinetochore attachment to the spindle occurs by a complex sequence of events. At the end of prophase in animal cells, the centrosomes of the growing spindle generally lie on opposite sides of the nuclear envelope. Thus, when the envelope breaks down, the sister-chromatid pairs are bombarded by microtubule plus ends coming from two directions. However, the kinetochores do not instantly achieve the correct ‘end-on’ microtubule attachment to both spindle poles. Instead, detailed studies with light and electron microscopy show that most initial attachments are unstable lateral attachments, in which a kinetochore attaches to the side of a passing microtubule, with assistance from kinesin motor proteins in the outer kinetochore. Soon, however, the dynamic microtubule plus ends capture the kinetochores in the correct end-on orientation
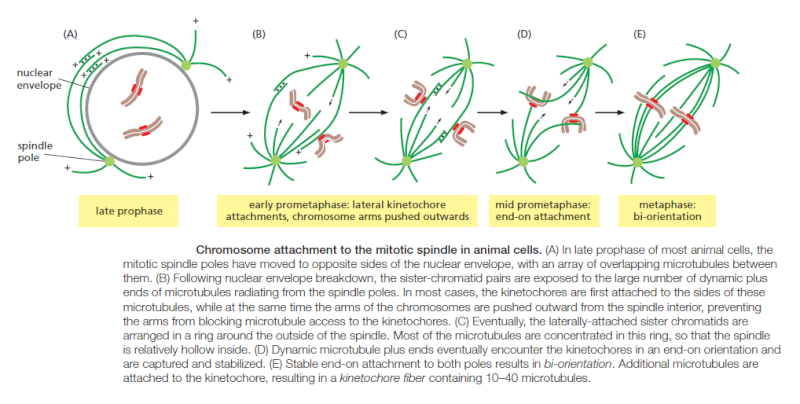
Another attachment mechanism also plays a part, particularly in the absence of centrosomes. Careful microscopic analysis suggests that short microtubules in the vicinity of the chromosomes become embedded in the plus-end-binding sites of the kinetochore. Polymerization at these plus ends then results in growth of the microtubules away from the kinetochore. The minus ends of these kinetochore microtubules are eventually cross-linked to other minus ends and focused by motor proteins at the spindle pole.
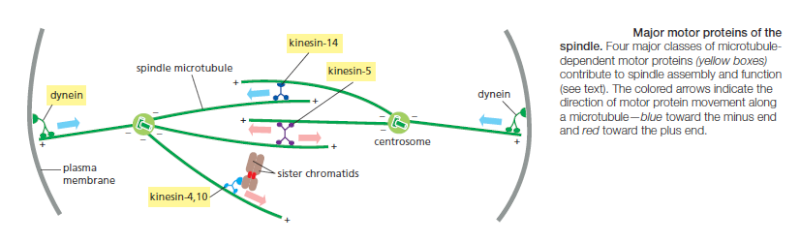
Bi-orientation is achieved by trial and error
The success of mitosis demands that sister chromatids in a pair attach to opposite poles of the mitotic spindle, so that they move to opposite ends of the cell when they separate in anaphase. How is this mode of attachment, called bi-orientation, achieved? What prevents the attachment of both kinetochores to the same spindle pole or the attachment of one kinetochore to both spindle poles? Part of the answer is that sister kinetochores are constructed in a back-to-back orientation that reduces the likelihood that both kinetochores can face the same spindle pole. Nevertheless, incorrect attachments do occur, and elegant regulatory mechanisms have evolved to correct them. Incorrect attachments are corrected by a system of trial and error that is based on a simple principle: incorrect attachments are highly unstable and do not last, whereas correct attachments become locked in place. How does the kinetochore sense a correct attachment? The answer appears to be tension .

When a sister-chromatid pair is properly bi-oriented on the spindle, the two kinetochores are pulled in opposite directions by strong poleward forces. Sister-chromatid cohesion resists these poleward forces, creating high levels of tension within the kinetochores. When chromosomes are incorrectly attached—when both sister chromatids are attached to the same spindle pole, for example—tension is low and the kinetochore sends an inhibitory signal that loosens the grip of its microtubule attachment site, allowing detachment to occur. When bi-orientation occurs, the high tension at the kinetochore shuts off the inhibitory signal, strengthening microtubule attachment. In animal cells, tension not only increases the affinity of the attachment site but also leads to the attachment of additional microtubules to the kinetochore. This results in the formation of a thick kinetochore fiber composed of multiple microtubules. The tension-sensing mechanism depends on the protein kinase Aurora-B, which is associated with the kinetochore and is thought to generate the inhibitory signal that reduces the strength of microtubule attachment in the absence of tension. It phosphorylates several components of the microtubule attachment site, including the Ndc80 complex, decreasing the site’s affinity for a microtubule plus end. When bi-orientation occurs, the resulting tension somehow reduces phosphorylation by Aurora-B, thereby increasing the affinity of the attachment site.
How tension might increase microtubule attachment to the kinetochore. ( see picture below ) These diagrams illustrate one speculative mechanism by which bi-orientation might increase microtubule attachment to the kinetochore. A single kinetochore is shown for clarity; the spindle pole is on the right. (A) When a sisterchromatid pair is unattached to the spindle or attached to just one spindle pole, there is little tension between the outer and inner kinetochores. The protein kinase Aurora-B is tethered to the inner kinetochore and phosphorylates the microtubule attachment sites, including the Ndc80 complex (blue), in the outer kinetochore as shown, thereby reducing the affinity of microtubule binding. Microtubules therefore associate and dissociate rapidly, and attachment is unstable. (B) When bi-orientation is achieved, the forces pulling the kinetochore toward the spindle pole are resisted by forces pulling the other sister kinetochore toward the opposite pole, and the resulting tension pulls the outer kinetochore away from the inner kinetochore. As a result, Aurora-B is unable to reach the outer kinetochore, and microtubule attachment sites are not phosphorylated. Microtubule binding affinity is therefore increased, resulting in the stable attachment of multiple microtubules to both kinetochores. The dephosphorylation of outer kinetochore proteins depends on a phosphatase that
is not shown here.
Following their attachment to the two spindle poles, the chromosomes are tugged back and forth, eventually assuming a position equidistant between the two poles, a position called the metaphase plate. In vertebrate cells, the chromosomes then oscillate gently at the metaphase plate, awaiting the signal for the sister chromatids to separate. The signal is produced, with a predictable lag time, after the bi-oriented attachment of the last of the chromosomes.Multiple Forces Act on Chromosomes in the Spindle
Multiple mechanisms generate the forces that move chromosomes back and forth after they are attached to the spindle, and produce the tension that is so important for the stabilization of correct attachments. In anaphase, similar forces pull the separated chromatids to opposite ends of the spindle. Three major spindle forces are particularly critical, although their strength and importance vary at different stages of mitosis. The first major force pulls the kinetochore and its associated chromatid along the kinetochore microtubule toward the spindle pole. It is produced by proteins at the kinetochore itself. By an uncertain mechanism, depolymerization at the plus end of the microtubule generates a force that pulls the kinetochore poleward. This force pulls on chromosomes during prometaphase and metaphase but is particularly important for moving sister chromatids toward the poles after they separate in anaphase. Interestingly, this kinetochore-generated poleward force does not require ATP or motor proteins. This might seem implausible at first, but it has been shown that purified kinetochores in a test tube, with no ATP present, can remain attached to depolymerizing microtubules and thereby move. The energy that drives the movement is stored in the microtubule and is released when the microtubule depolymerizes; it ultimately comes from the hydrolysis of GTP that occurs after a tubulin subunit adds to the end of a microtubule
How does plus-end depolymerization drive the kinetochore toward the pole? Ndc80 complexes in the kinetochore make multiple low-affinity attachments along the side of the microtubule. Because the attachments are constantly breaking and re-forming at new sites, the kinetochore remains attached to a microtubule even as the microtubule depolymerizes. In principle, this could move the kinetochore toward the spindle pole. A second poleward force is provided in some cell types by microtubule flux, whereby the microtubules themselves are pulled toward the spindle poles and dismantled at their minus ends. The mechanism underlying this poleward movement is not clear, although it might depend on forces generated by motor proteins and minus-end depolymerization at the spindle pole. In metaphase, the addition of new tubulin at the plus end of a microtubule compensates for the loss of tubulin at the minus end, so that microtubule length remains constant despite the movement of microtubules toward the spindle pole
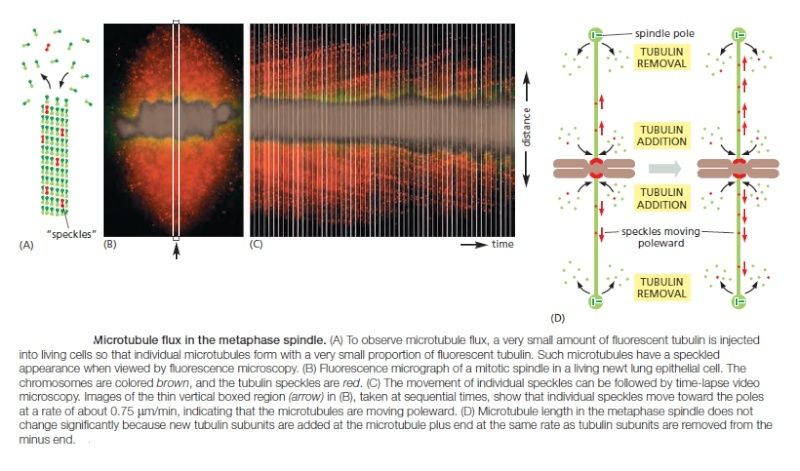
Any kinetochore that is attached to a microtubule undergoing such flux experiences a poleward force, which contributes to the generation of tension at the kinetochore in metaphase. Together with the kinetochore-based forces discussed above, flux also contributes to the poleward forces that move sister chromatids after they separate in anaphase.
The APC/C Triggers Sister-Chromatid Separation and the Completion of Mitosis
After M-Cdk has triggered the complex processes leading up to metaphase, the cell cycle reaches its climax with the separation of the sister chromatids at the metaphase-to-anaphase transition

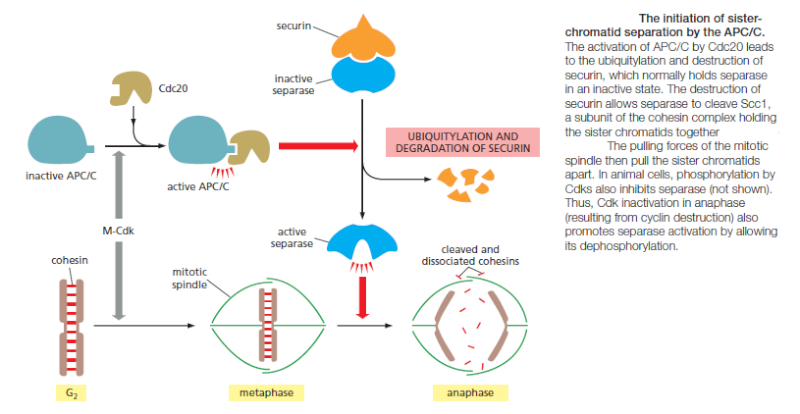
Although M-Cdk activity sets the stage for this event, the anaphase-promoting complex (APC/C) throws the switch that initiates sister-chromatid separation by ubiquitylating several mitotic regulatory proteins and thereby triggering their destruction During metaphase, cohesins holding the sister chromatids together resist the poleward forces that pull the sister chromatids apart. Anaphase begins with the sudden loss of sister-chromatid cohesion, which allows the sisters to separate and move to opposite poles of the spindle. The APC/C initiates the process by targeting the inhibitory protein securin for destruction. Before anaphase, securin binds to and inhibits the activity of a protease called separase. The destruction of securin at the end of metaphase releases separase, which is then free to cleave one of the subunits of cohesin. The cohesins fall away, and the sister chromatids
separate.
In addition to securin, the APC/C also targets the S- and M-cyclins for destruction, leading to the loss of most Cdk activity in anaphase. Cdk inactivation allows phosphatases to dephosphorylate the many Cdk target substrates in the cell, as required for the completion of mitosis and cytokinesis. If the APC/C triggers anaphase, what activates the APC/C? The answer is only partly known. As mentioned earlier, APC/C activation requires binding to the protein Cdc20. At least two processes regulate Cdc20 and its association with the APC/C. First, Cdc20 synthesis increases as the cell approaches mitosis, owing to an increase in the transcription of its gene. Second, phosphorylation of the APC/C helps Cdc20 bind to the APC/C, thereby helping to create an active complex. Among the kinases that phosphorylate and thus activate the APC/C is M-Cdk. Thus, M-Cdk not only triggers the early mitotic events leading up to metaphase, but it also sets the stage for progression into anaphase. The ability of M-Cdk to promote Cdc20–APC/C activity creates a negative feedback loop: M-Cdk sets in motion a regulatory process that leads to cyclin destruction and thus its own inactivation.
Unattached Chromosomes Block Sister-Chromatid Separation:
The Spindle Assembly Checkpoint
A spindle assembly checkpoint mechanism ensures that cells do not enter anaphase until all chromosomes are correctly bi-oriented on the mitotic spindle. The spindle assembly checkpoint depends on a sensor mechanism that monitors the strength of microtubule attachment at the kinetochore, possibly by sensing tension. Any kinetochore that is not properly attached to the spindle sends out a diffusible negative signal that blocks Cdc20–APC/C activation throughout the cell
and thus blocks the metaphase-to-anaphase transition. When the last sister-chromatid pair is properly bi-oriented, this block is removed, allowing sister-chromatid
separation to occur.
The negative checkpoint signal depends on several proteins, including Mad2, which are recruited to unattached kinetochores . Detailed structural analyses of Mad2 suggest that the unattached kinetochore acts like an enzyme that catalyzes a change in the conformation of Mad2, so that Mad2, together with other proteins, can bind and inhibit Cdc20–APC/C. In mammalian somatic cells, the spindle assembly checkpoint determines the normal timing of anaphase. The destruction of securin in these cells begins moments after the last sister-chromatid pair becomes bi-oriented on the spindle, and anaphase begins about 20 minutes later. Experimental inhibition of the checkpoint mechanism causes premature sister-chromatid separation and anaphase. Surprisingly, the normal timing of anaphase does not depend on the spindle assembly checkpoint in some cells, such as yeasts and the cells of early frog and fly embryos. Other mechanisms, as yet unknown, must determine the timing of anaphase in these cells.
Chromosomes Segregate in Anaphase A and B
The sudden loss of sister-chromatid cohesion at the onset of anaphase leads to sister-chromatid separation, which allows the forces of the mitotic spindle to pull the sisters to opposite poles of the cell—called chromosome segregation. The chromosomes move by two independent and overlapping processes. The first, anaphase A, is the initial poleward movement of the chromosomes, which is accompanied by shortening of the kinetochore microtubules. The second, anaphase B, is the separation of the spindle poles themselves, which begins after the sister chromatids have separated and the daughter chromosomes have moved some distance apart.

Chromosome movement in anaphase A depends on a combination of the two major poleward forces described earlier. The first is the force generated by microtubule depolymerization at the kinetochore, which results in the loss of tubulin subunits at the plus end as the kinetochore moves toward the pole. The second is provided by microtubule flux, which is the poleward movement of the microtubules toward the spindle pole, where minus-end depolymerization occurs. The relative importance of these two forces during anaphase varies in different cell types: in embryonic cells, chromosome movement depends mainly on microtubule flux, for example, whereas movement in yeast and vertebrate somatic cells results primarily from forces generated at the kinetochore. Spindle-pole separation during anaphase B depends on motor driven mechanisms similar to those that separate the two centrosomes in early mitosis. Plusend directed kinesin-5 motor proteins, which cross-link the overlapping plus ends of the interpolar microtubules, push the poles apart. In addition, dynein motors that anchor astral microtubule plus ends to the cell cortex pull the poles apart.
Although sister-chromatid separation initiates the chromosome movements of anaphase A, other mechanisms also ensure correct chromosome movements in anaphase A and spindle elongation in anaphase B. Most importantly, the completion of a normal anaphase depends on the dephosphorylation of Cdk substrates, which in most cells results from the APC/C-dependent destruction of cyclins. If M-cyclin destruction is prevented—by the production of a mutant form that is not recognized by the APC/C, for example—sister-chromatid separation generally occurs, but the chromosome movements and microtubule behavior of anaphase are abnormal. The relative contributions of anaphase A and anaphase B to chromosome segregation vary greatly, depending on the cell type. In mammalian cells, anaphase B begins shortly after anaphase A and stops when the spindle is about twice its metaphase length; in contrast, the spindles of yeasts and certain protozoa primarily use anaphase B to separate the chromosomes at anaphase, and their spindles elongate to up to 15 times their metaphase length.
Segregated Chromosomes Are Packaged in Daughter Nuclei at Telophase
By the end of anaphase, the daughter chromosomes have segregated into two equal groups at opposite ends of the cell. In telophase, the final stage of mitosis, the two sets of chromosomes are packaged into a pair of daughter nuclei. The first major event of telophase is the disassembly of the mitotic spindle, followed by the re-formation of the nuclear envelope. Initially, nuclear membrane fragments associate with the surface of individual chromosomes. These membrane fragments fuse to partly enclose clusters of chromosomes and then coalesce to reform the complete nuclear envelope. Nuclear pore complexes are incorporated into the envelope, the nuclear lamina re-forms, and the envelope once again becomes continuous with the endoplasmic reticulum. Once the nuclear envelope has re-formed, the pore complexes pump in nuclear proteins, the nucleus expands, and the mitotic chromosomes are reorganized into their interphase state, allowing gene transcription to resume. A new nucleus has been created, and mitosis is complete. All that remains is for the cell to complete its division into two. We saw earlier that phosphorylation of various proteins by M-Cdk promotes spindle assembly, chromosome condensation, and nuclear-envelope breakdown in early mitosis. It is thus not surprising that the dephosphorylation of these same proteins is required for spindle disassembly and the re-formation of daughter nuclei in telophase. In principle, these dephosphorylations and the completion of mitosis could be triggered by the inactivation of Cdks, the activation of phosphatases, or both. Although Cdk inactivation—resulting primarily from cyclin destruction—is mainly responsible in most cells, some cells also rely on activation of phosphatases. In budding yeast, for example, the completion of mitosis depends on the activation of a phosphatase called Cdc14, which dephosphorylates
a subset of Cdk substrates involved in anaphase and telophase.
Summary
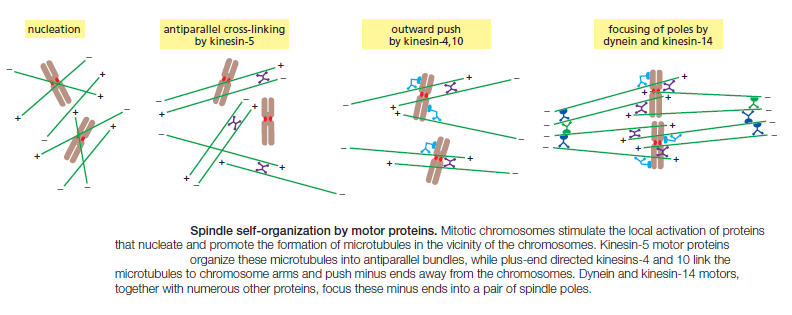
M-Cdk triggers the events of early mitosis, including chromosome condensation, assembly of the mitotic spindle, and bipolar attachment of the sister-chromatid pairs to microtubules of the spindle. Spindle formation in animal cells depends largely on the ability of mitotic chromosomes to stimulate local microtubule nucleation and stability, as well as on the ability of motor proteins to organize microtubules into a bipolar array. Many cells also use centrosomes to facilitate spindle assembly. Anaphase is triggered by the APC/C, which stimulates the destruction of the proteins that hold the sister chromatids together. APC/C also promotes cyclin destruction and thus the inactivation of M-Cdk. The resulting dephosphorylation of Cdk targets is required for the events that complete mitosis, including the disassembly of the spindle and the re-formation of the nuclear envelope.
The kinetochore is a network of protein complexes that assembles on centromere regions of chromatin and acts as the connection point between chromatids and the spindle microtubules that segregate them into daughter cells. Kinetochores must sense microtubule attachment and determine whether this attachment of sister chromatids is to the same or different spindle poles. The stable engagement of a kinetochore is regulated by the Aurora B kinase, the activity of which has been shown to reduce the affinity of several kinetochore proteins for microtubules. A single unattached or incorrectly attached kinetochore is sufficient to trigger the spindle assembly checkpoint and halt progress into anaphase, preventing cell division. 1 Once all of the chromosomes in a cell preparing to divide are correctly bioriented and satisfy the spindle assembly checkpoint, chromosomes must be pulled apart into daughter cells. An essential function of the kinetochore is to couple chromosome movement to microtubule depolymerization. Kinetochores are able to track depolymerizing microtubule ends and harness the energy released during microtubule depolymerization to move chromosomes to opposite spindle poles.
Kinetochores attach sister chromatids to the Spindle
Following the assembly of a bipolar microtubule array, the second major step in spindle formation is the attachment of the array to the sister-chromatid pairs. Spindle microtubules become attached to each chromatid at its kinetochore, a giant, multilayered protein structure that is built at the centromeric region of the chromatid (see picture below)
The kinetochore. see below (A) A fluorescence micrograph of a metaphase chromosome stained with a DNA-binding fluorescent dye and with human autoantibodies that react with specific kinetochore proteins. The two kinetochores, one associated with each sister chromatid, are stained red. (B) A drawing of a metaphase chromosome showing its two sister chromatids attached to the plus ends of kinetochore microtubules. Each kinetochore forms a plaque on the surface of the centromere. (C) Electron micrograph of an anaphase chromatid with microtubules attached to its kinetochore. While most kinetochores have a trilaminar structure, the one shown here (from a green alga) has an unusually complex structure with additional layers.

In metaphase, the plus ends of kinetochore microtubules are embedded head-on in specialized microtubuleattachment sites within the outer region of the kinetochore, furthest from the DNA. The kinetochore of an animal cell can bind 10–40 microtubules, whereas a budding yeast kinetochore can bind only one. Attachment of each microtubule depends on multiple copies of a rod-shaped protein complex called the Ndc80 complex, which is anchored in the kinetochore at one end and interacts with the sides of the microtubule at the other, thereby linking the microtubule to the kinetochore while still allowing the addition or removal of tubulin subunits at this end.

Regulation of plus-end polymerization and depolymerization at the kinetochore is critical for the control of chromosome movement on the spindle. Kinetochore attachment to the spindle occurs by a complex sequence of events. At the end of prophase in animal cells, the centrosomes of the growing spindle generally lie on opposite sides of the nuclear envelope. Thus, when the envelope breaks down, the sister-chromatid pairs are bombarded by microtubule plus ends coming from two directions. However, the kinetochores do not instantly achieve the correct ‘end-on’ microtubule attachment to both spindle poles. Instead, detailed studies with light and electron microscopy show that most initial attachments are unstable lateral attachments, in which a kinetochore attaches to the side of a passing microtubule, with assistance from kinesin motor proteins in the outer kinetochore. Soon, however, the dynamic microtubule plus ends capture the kinetochores in the correct end-on orientation

Another attachment mechanism also plays a part, particularly in the absence of centrosomes. Careful microscopic analysis suggests that short microtubules in the vicinity of the chromosomes become embedded in the plus-end-binding sites of the kinetochore. Polymerization at these plus ends then results in growth of the microtubules away from the kinetochore. The minus ends of these kinetochore microtubules are eventually cross-linked to other minus ends and focused by motor proteins at the spindle pole.


Bi-orientation is achieved by trial and error
The success of mitosis demands that sister chromatids in a pair attach to opposite poles of the mitotic spindle, so that they move to opposite ends of the cell when they separate in anaphase. How is this mode of attachment, called bi-orientation, achieved? What prevents the attachment of both kinetochores to the same spindle pole or the attachment of one kinetochore to both spindle poles? Part of the answer is that sister kinetochores are constructed in a back-to-back orientation that reduces the likelihood that both kinetochores can face the same spindle pole. Nevertheless, incorrect attachments do occur, and elegant regulatory mechanisms have evolved to correct them. Incorrect attachments are corrected by a system of trial and error that is based on a simple principle: incorrect attachments are highly unstable and do not last, whereas correct attachments become locked in place. How does the kinetochore sense a correct attachment? The answer appears to be tension .

When a sister-chromatid pair is properly bi-oriented on the spindle, the two kinetochores are pulled in opposite directions by strong poleward forces. Sister-chromatid cohesion resists these poleward forces, creating high levels of tension within the kinetochores. When chromosomes are incorrectly attached—when both sister chromatids are attached to the same spindle pole, for example—tension is low and the kinetochore sends an inhibitory signal that loosens the grip of its microtubule attachment site, allowing detachment to occur. When bi-orientation occurs, the high tension at the kinetochore shuts off the inhibitory signal, strengthening microtubule attachment. In animal cells, tension not only increases the affinity of the attachment site but also leads to the attachment of additional microtubules to the kinetochore. This results in the formation of a thick kinetochore fiber composed of multiple microtubules. The tension-sensing mechanism depends on the protein kinase Aurora-B, which is associated with the kinetochore and is thought to generate the inhibitory signal that reduces the strength of microtubule attachment in the absence of tension. It phosphorylates several components of the microtubule attachment site, including the Ndc80 complex, decreasing the site’s affinity for a microtubule plus end. When bi-orientation occurs, the resulting tension somehow reduces phosphorylation by Aurora-B, thereby increasing the affinity of the attachment site.
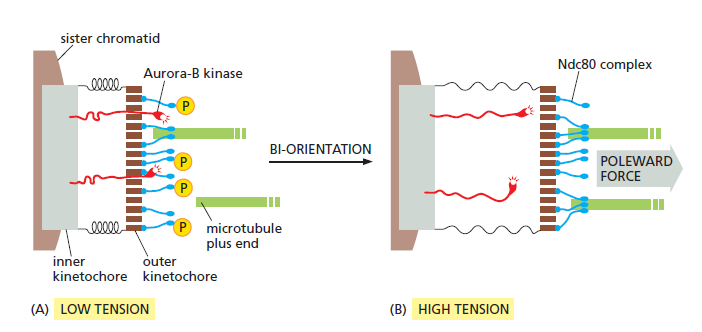
How tension might increase microtubule attachment to the kinetochore. ( see picture below ) These diagrams illustrate one speculative mechanism by which bi-orientation might increase microtubule attachment to the kinetochore. A single kinetochore is shown for clarity; the spindle pole is on the right. (A) When a sisterchromatid pair is unattached to the spindle or attached to just one spindle pole, there is little tension between the outer and inner kinetochores. The protein kinase Aurora-B is tethered to the inner kinetochore and phosphorylates the microtubule attachment sites, including the Ndc80 complex (blue), in the outer kinetochore as shown, thereby reducing the affinity of microtubule binding. Microtubules therefore associate and dissociate rapidly, and attachment is unstable. (B) When bi-orientation is achieved, the forces pulling the kinetochore toward the spindle pole are resisted by forces pulling the other sister kinetochore toward the opposite pole, and the resulting tension pulls the outer kinetochore away from the inner kinetochore. As a result, Aurora-B is unable to reach the outer kinetochore, and microtubule attachment sites are not phosphorylated. Microtubule binding affinity is therefore increased, resulting in the stable attachment of multiple microtubules to both kinetochores. The dephosphorylation of outer kinetochore proteins depends on a phosphatase that
is not shown here.

Following their attachment to the two spindle poles, the chromosomes are tugged back and forth, eventually assuming a position equidistant between the two poles, a position called the metaphase plate. In vertebrate cells, the chromosomes then oscillate gently at the metaphase plate, awaiting the signal for the sister chromatids to separate. The signal is produced, with a predictable lag time, after the bi-oriented attachment of the last of the chromosomes.
Multiple mechanisms generate the forces that move chromosomes back and forth after they are attached to the spindle, and produce the tension that is so important for the stabilization of correct attachments. In anaphase, similar forces pull the separated chromatids to opposite ends of the spindle. Three major spindle forces are particularly critical, although their strength and importance vary at different stages of mitosis. The first major force pulls the kinetochore and its associated chromatid along the kinetochore microtubule toward the spindle pole. It is produced by proteins at the kinetochore itself. By an uncertain mechanism, depolymerization at the plus end of the microtubule generates a force that pulls the kinetochore poleward. This force pulls on chromosomes during prometaphase and metaphase but is particularly important for moving sister chromatids toward the poles after they separate in anaphase. Interestingly, this kinetochore-generated poleward force does not require ATP or motor proteins. This might seem implausible at first, but it has been shown that purified kinetochores in a test tube, with no ATP present, can remain attached to depolymerizing microtubules and thereby move. The energy that drives the movement is stored in the microtubule and is released when the microtubule depolymerizes; it ultimately comes from the hydrolysis of GTP that occurs after a tubulin subunit adds to the end of a microtubule
How does plus-end depolymerization drive the kinetochore toward the pole? Ndc80 complexes in the kinetochore make multiple low-affinity attachments along the side of the microtubule. Because the attachments are constantly breaking and re-forming at new sites, the kinetochore remains attached to a microtubule even as the microtubule depolymerizes. In principle, this could move the kinetochore toward the spindle pole. A second poleward force is provided in some cell types by microtubule flux, whereby the microtubules themselves are pulled toward the spindle poles and dismantled at their minus ends. The mechanism underlying this poleward movement is not clear, although it might depend on forces generated by motor proteins and minus-end depolymerization at the spindle pole. In metaphase, the addition of new tubulin at the plus end of a microtubule compensates for the loss of tubulin at the minus end, so that microtubule length remains constant despite the movement of microtubules toward the spindle pole

Any kinetochore that is attached to a microtubule undergoing such flux experiences a poleward force, which contributes to the generation of tension at the kinetochore in metaphase. Together with the kinetochore-based forces discussed above, flux also contributes to the poleward forces that move sister chromatids after they separate in anaphase.
The APC/C Triggers Sister-Chromatid Separation and the Completion of Mitosis
After M-Cdk has triggered the complex processes leading up to metaphase, the cell cycle reaches its climax with the separation of the sister chromatids at the metaphase-to-anaphase transition

Although M-Cdk activity sets the stage for this event, the anaphase-promoting complex (APC/C) throws the switch that initiates sister-chromatid separation by ubiquitylating several mitotic regulatory proteins and thereby triggering their destruction During metaphase, cohesins holding the sister chromatids together resist the poleward forces that pull the sister chromatids apart. Anaphase begins with the sudden loss of sister-chromatid cohesion, which allows the sisters to separate and move to opposite poles of the spindle. The APC/C initiates the process by targeting the inhibitory protein securin for destruction. Before anaphase, securin binds to and inhibits the activity of a protease called separase. The destruction of securin at the end of metaphase releases separase, which is then free to cleave one of the subunits of cohesin. The cohesins fall away, and the sister chromatids
separate.


In addition to securin, the APC/C also targets the S- and M-cyclins for destruction, leading to the loss of most Cdk activity in anaphase. Cdk inactivation allows phosphatases to dephosphorylate the many Cdk target substrates in the cell, as required for the completion of mitosis and cytokinesis. If the APC/C triggers anaphase, what activates the APC/C? The answer is only partly known. As mentioned earlier, APC/C activation requires binding to the protein Cdc20. At least two processes regulate Cdc20 and its association with the APC/C. First, Cdc20 synthesis increases as the cell approaches mitosis, owing to an increase in the transcription of its gene. Second, phosphorylation of the APC/C helps Cdc20 bind to the APC/C, thereby helping to create an active complex. Among the kinases that phosphorylate and thus activate the APC/C is M-Cdk. Thus, M-Cdk not only triggers the early mitotic events leading up to metaphase, but it also sets the stage for progression into anaphase. The ability of M-Cdk to promote Cdc20–APC/C activity creates a negative feedback loop: M-Cdk sets in motion a regulatory process that leads to cyclin destruction and thus its own inactivation.
Unattached Chromosomes Block Sister-Chromatid Separation:
The Spindle Assembly Checkpoint
A spindle assembly checkpoint mechanism ensures that cells do not enter anaphase until all chromosomes are correctly bi-oriented on the mitotic spindle. The spindle assembly checkpoint depends on a sensor mechanism that monitors the strength of microtubule attachment at the kinetochore, possibly by sensing tension. Any kinetochore that is not properly attached to the spindle sends out a diffusible negative signal that blocks Cdc20–APC/C activation throughout the cell
and thus blocks the metaphase-to-anaphase transition. When the last sister-chromatid pair is properly bi-oriented, this block is removed, allowing sister-chromatid
separation to occur.
The negative checkpoint signal depends on several proteins, including Mad2, which are recruited to unattached kinetochores . Detailed structural analyses of Mad2 suggest that the unattached kinetochore acts like an enzyme that catalyzes a change in the conformation of Mad2, so that Mad2, together with other proteins, can bind and inhibit Cdc20–APC/C. In mammalian somatic cells, the spindle assembly checkpoint determines the normal timing of anaphase. The destruction of securin in these cells begins moments after the last sister-chromatid pair becomes bi-oriented on the spindle, and anaphase begins about 20 minutes later. Experimental inhibition of the checkpoint mechanism causes premature sister-chromatid separation and anaphase. Surprisingly, the normal timing of anaphase does not depend on the spindle assembly checkpoint in some cells, such as yeasts and the cells of early frog and fly embryos. Other mechanisms, as yet unknown, must determine the timing of anaphase in these cells.
Chromosomes Segregate in Anaphase A and B
The sudden loss of sister-chromatid cohesion at the onset of anaphase leads to sister-chromatid separation, which allows the forces of the mitotic spindle to pull the sisters to opposite poles of the cell—called chromosome segregation. The chromosomes move by two independent and overlapping processes. The first, anaphase A, is the initial poleward movement of the chromosomes, which is accompanied by shortening of the kinetochore microtubules. The second, anaphase B, is the separation of the spindle poles themselves, which begins after the sister chromatids have separated and the daughter chromosomes have moved some distance apart.

Chromosome movement in anaphase A depends on a combination of the two major poleward forces described earlier. The first is the force generated by microtubule depolymerization at the kinetochore, which results in the loss of tubulin subunits at the plus end as the kinetochore moves toward the pole. The second is provided by microtubule flux, which is the poleward movement of the microtubules toward the spindle pole, where minus-end depolymerization occurs. The relative importance of these two forces during anaphase varies in different cell types: in embryonic cells, chromosome movement depends mainly on microtubule flux, for example, whereas movement in yeast and vertebrate somatic cells results primarily from forces generated at the kinetochore. Spindle-pole separation during anaphase B depends on motor driven mechanisms similar to those that separate the two centrosomes in early mitosis. Plusend directed kinesin-5 motor proteins, which cross-link the overlapping plus ends of the interpolar microtubules, push the poles apart. In addition, dynein motors that anchor astral microtubule plus ends to the cell cortex pull the poles apart.
Although sister-chromatid separation initiates the chromosome movements of anaphase A, other mechanisms also ensure correct chromosome movements in anaphase A and spindle elongation in anaphase B. Most importantly, the completion of a normal anaphase depends on the dephosphorylation of Cdk substrates, which in most cells results from the APC/C-dependent destruction of cyclins. If M-cyclin destruction is prevented—by the production of a mutant form that is not recognized by the APC/C, for example—sister-chromatid separation generally occurs, but the chromosome movements and microtubule behavior of anaphase are abnormal. The relative contributions of anaphase A and anaphase B to chromosome segregation vary greatly, depending on the cell type. In mammalian cells, anaphase B begins shortly after anaphase A and stops when the spindle is about twice its metaphase length; in contrast, the spindles of yeasts and certain protozoa primarily use anaphase B to separate the chromosomes at anaphase, and their spindles elongate to up to 15 times their metaphase length.
Segregated Chromosomes Are Packaged in Daughter Nuclei at Telophase
By the end of anaphase, the daughter chromosomes have segregated into two equal groups at opposite ends of the cell. In telophase, the final stage of mitosis, the two sets of chromosomes are packaged into a pair of daughter nuclei. The first major event of telophase is the disassembly of the mitotic spindle, followed by the re-formation of the nuclear envelope. Initially, nuclear membrane fragments associate with the surface of individual chromosomes. These membrane fragments fuse to partly enclose clusters of chromosomes and then coalesce to reform the complete nuclear envelope. Nuclear pore complexes are incorporated into the envelope, the nuclear lamina re-forms, and the envelope once again becomes continuous with the endoplasmic reticulum. Once the nuclear envelope has re-formed, the pore complexes pump in nuclear proteins, the nucleus expands, and the mitotic chromosomes are reorganized into their interphase state, allowing gene transcription to resume. A new nucleus has been created, and mitosis is complete. All that remains is for the cell to complete its division into two. We saw earlier that phosphorylation of various proteins by M-Cdk promotes spindle assembly, chromosome condensation, and nuclear-envelope breakdown in early mitosis. It is thus not surprising that the dephosphorylation of these same proteins is required for spindle disassembly and the re-formation of daughter nuclei in telophase. In principle, these dephosphorylations and the completion of mitosis could be triggered by the inactivation of Cdks, the activation of phosphatases, or both. Although Cdk inactivation—resulting primarily from cyclin destruction—is mainly responsible in most cells, some cells also rely on activation of phosphatases. In budding yeast, for example, the completion of mitosis depends on the activation of a phosphatase called Cdc14, which dephosphorylates
a subset of Cdk substrates involved in anaphase and telophase.
Summary
M-Cdk triggers the events of early mitosis, including chromosome condensation, assembly of the mitotic spindle, and bipolar attachment of the sister-chromatid pairs to microtubules of the spindle. Spindle formation in animal cells depends largely on the ability of mitotic chromosomes to stimulate local microtubule nucleation and stability, as well as on the ability of motor proteins to organize microtubules into a bipolar array. Many cells also use centrosomes to facilitate spindle assembly. Anaphase is triggered by the APC/C, which stimulates the destruction of the proteins that hold the sister chromatids together. APC/C also promotes cyclin destruction and thus the inactivation of M-Cdk. The resulting dephosphorylation of Cdk targets is required for the events that complete mitosis, including the disassembly of the spindle and the re-formation of the nuclear envelope.
Last edited by Admin on Tue Jul 21, 2015 12:09 pm; edited 16 times in total




