http://reasonandscience.heavenforum.org/t2029-ribonucleotide-reductase-one-of-the-most-essential-enzymes-of-life-and-how-it-buries-the-rna-world#3428
This is one of the most essential enzymes of life 5
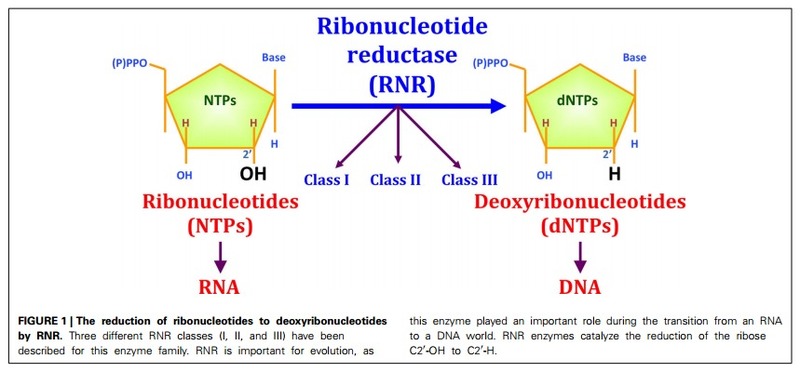
Ribonucleotide reduction is the only pathway for de novo synthesis of deoxyribonucleotides in extant organisms. This chemically demanding reaction, which proceeds via a carbon-centered free radical, is catalyzed by ribonucleotide reductase (RNR). The mechanism has been deemed unlikely to be catalyzed by a ribozyme, creating an enigma regarding how the building blocks for DNA were synthesized at the transition from RNA to DNA-encoded genomes.
DNA is the genetic material in all cellular organisms plus many viruses. DNA’s building blocks, deoxyribonucleotides (dNTPs), are always synthesized by reduction of ribonucleotides (either NTPs or NDPs), the building blocks of RNA. 6
Origin of Ribonucleotide Reduction
How and when ribonucleotide reduction evolved is a question that is intimately associated with the transition from the RNA world to the modern RNA + protein + DNA world, since it is the only known de novo mechanism for dNTP synthesis.
The maintenance of life on Earth depends on the ability to reproduce. Reproduction requires an accurate and stable storage system for the genetic information in all organisms, including viruses. It has been recently suggested that the RNA molecule, with autoreplicative capacity, is the primary primitive molecule for the genetic information storage. Despite the wide acceptance of this idea, there are arguments against the concept of an RNA world that cannot be underestimated. 7
Today, three different RNR classes have been described, with little apparent similarity between them in terms of primary protein sequence (approximately 10–20% similarity). Thus, it could be assumed that each RNR class appeared independently from each other over time.
There we have a problem of convergent evolution. “ As Stephen J.Gould stated :
…No finale can be specified at the start, none would ever occur a second time in the same way, because any pathway proceeds through thousands of improbable stages. Alter any early event, ever so slightly, and without apparent importance at the time, and evolution cascades into a radically different channel. 11
That means, hardly we should find a enzyme evolving the same function. But thats exactly what supposedly happened. Not only did the RNR would have had to arise 3 times independently with different gene sets, but provided the same function. Should we not expect it to evolve just once, if the function is the same ?
photos upload
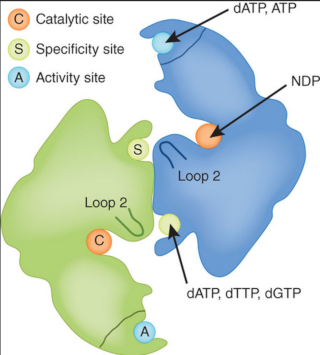
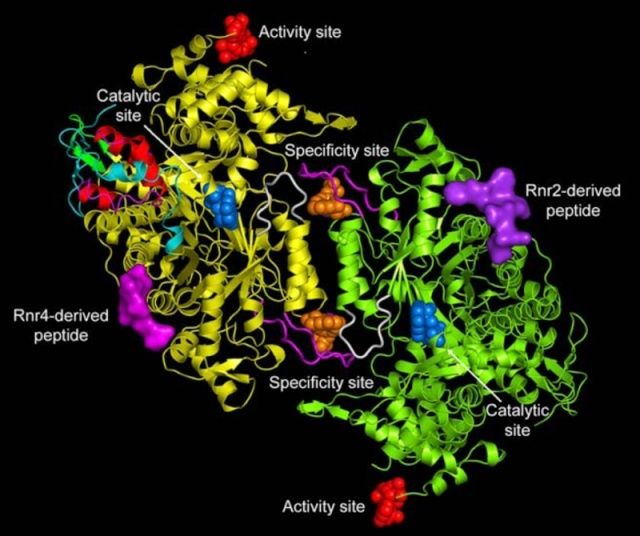
Three main classes of ribonucleotide reductases (RNR) have been discovered that depend on different metal cofactors for the catalytic activity:
class I enzymes contain a diiron-oxygen cluster,
class II a cobalt containing cobalamin cofactor (vitamin B12), and
class III an 4Fe-4S iron-sulfur cluster coupled to S-adenosylmethionine (SAM) [PMID: 15158709].
But, surprisingly, there is a great similarity the reaction mechanism, allosteric regulation and three-dimensional structure (tertiary structure) of these enzymes, suggesting a potential common origin.
The enzymatic activation of class III RNR requires Sadenosylmethionine (SAM), one of the most ancestral molecules, with few steps required for its biosynthesis
Biosynthesis DNA is made from RNA. The deoxynucleotides are made from nucleotides with ribonucleotide reductases (RNR's), producing uracil-DNA or u-DNA. The uracil is then converted to thymine by adding a methyl group, making thymine-DNA or t-DNA, the kind that is actually used. 4)
The reaction catalyzed by RNR is strictly conserved in all living organisms. Furthermore RNR plays a critical role in regulating the total rate of DNA synthesis so that DNA to cell mass is maintained at a constant ratio during cell division and DNA repair. A somewhat unusual feature of the RNR enzyme is that it catalyzes a reaction that proceeds via a free radical mechanism of action.The substrates for RNR are ADP, GDP, CDP and UDP. dTDP (deoxythymidine diphosphate) is synthesized by another enzyme (thymidylate kinase) from dTMP (deoxythymidine monophosphate). 1
The iron-dependent enzyme, ribonucleotide reductase (RNR), is essential for DNA synthesis.
The structures of a class III ribonucleotide reductase (RNR) and pyruvate formate lyase exhibit striking homology within their active site domains with respect to each other and to the previously published structure of a class I RNR. The common structures and the common complex-radical-based chemistry of these systems, as well as of the class II RNRs, suggest that RNRs evolved by divergent evolution and provide an essential link between the RNA and DNA world. 2
Genetic information can be stored stably only because a battery of DNA repair enzymes scan the DNA and replace the damaged nucleotides. Without these enzymes it would be inconceivable how primitive cells kept abreast of the constant high-level damage by the environment and by endogenous reactions. If unrepaired, cell death would result. 10
RNR is a complex of two dimeric proteins termed R1 and R2. 8
That brings us to the classic chicken and egg, catch22 situation. RNR enzymes are required to make DNA. DNA is however required to make RNR enzymes. What came first ??
We can conclude with high certainty that this enzyme buries any RNA world fantasies, and any possibility of transition from RNA to DNA world scenarios.

windows screen capture

how to do a screenshot on a pc 9

1) http://en.wikipedia.org/wiki/Ribonucleotide_reductase
2) http://journal.frontiersin.org/article/10.3389/fcimb.2014.00052/abstract
3) http://www.ncbi.nlm.nih.gov/pubmed/11114511
4) http://evolutionwiki.org/wiki/RNA_world
5) http://www.nature.com/nsmb/journal/v18/n3/full/nsmb0311-251.html
6) file:///E:/Downloads/life-05-00604-v2.pdf
7) file:///E:/Downloads/fcimb-04-00052.pdf
8 ) http://ac.els-cdn.com/S0969212696001128/1-s2.0-S0969212696001128-main.pdf?_tid=c961a940-0289-11e5-82eb-00000aacb35f&acdnat=1432522831_8571d89fe458862e512f4f821993f918
9 ) http://www.mdpi.com/2075-1729/5/1/604/htm
10 ) http://creation.com/origin-of-life-critique#endRef49
11) Stephen J. Gould, Wonderful Life: The Burgess Shale and the Nature of History (New York, NY: W.W. Norton & Company, 1989)
further readings:
http://www.ncbi.nlm.nih.gov/pmc/articles/PMC19537/
Last edited by Admin on Fri 29 Jan 2016 - 10:45; edited 2 times in total





 9
9