Proteasome Garbage Grinders, evidence of luck, evolution, or design?
https://reasonandscience.catsboard.com/t1851-proteasome-garbage-grinders
Proteasome:
1. The disposal of protein “trash” in the cell is the job of a complex machine called the proteasome. What could be more low than trash collection? Here also, sophisticated mechanisms work together.
2. PhysOrg described a new finding that shows that “two different mechanisms are required to determine which targets to destroy.” The “recognition tag” and “initiator tag.”
3. Both mechanisms have to be aligned properly to enter the machine’s disposal barrel. “The proteasome can recognize different plugs1, but each one has to have the correct specific arrangement of prongs1,” said a researcher at Northwestern University.
4. This is another example of interdependent irreducible complex systems. One can’t argue for evolution; that first only one system existed.
5. The work of a designer is again obvious and all men call him God.
6. God most probably exists.
https://www.youtube.com/watch?v=hvNJ3yWZQbE&t=90s
http://iaincarstairs.wordpress.com/2013/03/25/as-smart-as-molecules/

Once proteins are formed in the cell, there are circumstances in which they need to be destroyed, for example, if incorrectly formed or of no further use. The cells have their own aggressive TSA agents ( Transportation Security Administration ) who spot and stamp such proteins with a ubiquitin molecule which alerts other transport systems that the protein needs to be scrapped.
http://www.nature.com/ncb/journal/v6/n11/full/ncb1104-1011.html
Selective protein degradation by the ubiquitin–proteasome pathway is a fundamental regulatory strategy that fulfils essential roles in various cellular processes.
http://www.nature.com/ncb/journal/v7/n8/full/ncb0805-725.html
Following the discovery of ubiquitin in the late 1970s and the delineation of the enzymology of the pathway, its regulatory potential for orchestrating complex cellular programmes was initially severely underestimated.
How did the first cell " know " that a recycle mechanism for damaged proteins would be useful ? How did it either evolve the garbage grinder, or how could it possibly have arisen by chance ( since it had to be present probably as a essential mechanism right in the beginning of the first cell ? )
The proteosome : This shredder has a twist-open lid which is activated by the unique combination of three destruction tags added to the errant protein.
https://www.youtube.com/watch?v=hvNJ3yWZQbE
http://en.wikipedia.org/wiki/Proteasome#cite_note-Haas-34
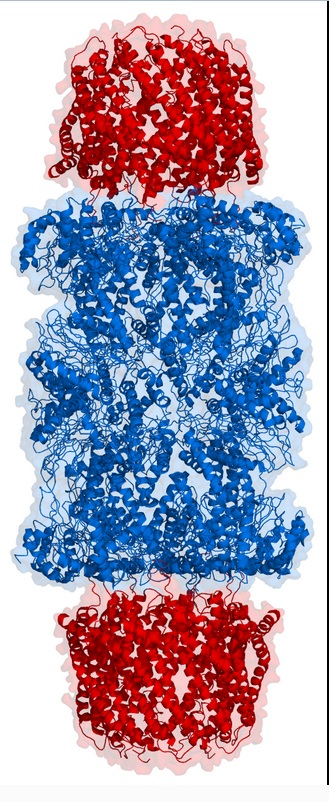
Proteasomes are protein complexes inside all eukaryotes and archaea, and in some bacteria. In eukaryotes, they are located in the nucleus and the cytoplasm.The main function of the proteasome is to degrade unneeded or damaged proteins by proteolysis, a chemical reaction that breaks peptide bonds
Enzymes that carry out such reactions are called
Proteasomes are part of a major mechanism by which cells regulate the concentration of particular proteins and degrade misfolded proteins. The degradation process yields peptides of about seven to eight amino acids long, which can then be further degraded into shorter amino acid sequences and used in synthesizing new proteins. Proteins are tagged for degradation with a small protein called ubiquitin.
How did ubiquitin arise ? What function would it have, if not fully inserted in the whole mechanism to do the job ?
The tagging reaction is catalyzed by enzymes called ubiquitin ligases
http://en.wikipedia.org/wiki/Ubiquitin_ligase
A ubiquitin ligase (also called an E3 ubiquitin ligase) is a ligase enzyme that combines with a ubiquitin-containing E2 ubiquitin-conjugating enzyme, recognizes the target protein that is to be ubiquinated, and causes the attachment of ubiquitin to a lysine on the target protein via an isopeptide bond. An E3 ubiquitin ligase targets specific protein substrates for degradation by the proteasome. In general, the ubiquitin ligase is involved in poly-ubiquitination: a second ubiquitin is attached to the first, a third is attached to the second, and so forth. Poly-ubiquitination marks proteins for degradation by the proteasome.
Once a protein is tagged with a single ubiquitin molecule, this is a signal to other ligases to attach additional ubiquitin molecules. The result is a polyubiquitin chain that is bound by the proteasome, allowing it to degrade the tagged protein.[2]
In structure, the proteasome is a cylindrical complex containing a "core" of four stacked rings forming a central pore.
How did this structure arise ? what survival advantage would there be for a unfinished structure ?
Each ring is composed of seven individual proteins. The inner two rings are made of seven β subunits that contain three to seven protease active sites. These sites are located on the interior surface of the rings, so that the target protein must enter the central pore before it is degraded. The outer two rings each contain seven α subunits whose function is to maintain a "gate" through which proteins enter the barrel.
How did the barrel shape arise ?
These α subunits are controlled by binding to "cap" structures or regulatory particles that recognize polyubiquitin tags attached to protein substrates and initiate the degradation process. The overall system of ubiquitination and proteasomal degradation is known as the ubiquitin-proteasome system.
The proteasomal degradation pathway is essential for many cellular processes, including the cell cycle, the regulation of gene expression, and responses to oxidative stress. The importance of proteolytic degradation inside cells and the role of ubiquitin in proteolytic pathways was acknowledged in the award of the 2004 Nobel Prize in Chemistry to Aaron Ciechanover, Avram Hershko and Irwin Rose.[3]
In the first step, a ubiquitin-activating enzyme (known as E1) hydrolyzes ATP and adenylylates a ubiquitin molecule.
http://en.wikipedia.org/wiki/Ubiquitin-activating_enzyme
Ubiquitin-activating enzymes, also known as E1 enzymes, catalyze the first step in the ubiquitination reaction, which (among other things) can target a protein for degradation via a proteasome. This covalent attachment of ubiquitin or ubiquitin-like proteins to targeted proteins is a major mechanism for regulating protein function in eukaryotic organisms.[2] Many processes such as cell division, immune responses and embryonic development are also regulated by post-transcriptional modification by ubiquitin and ubiquitin-like proteins.
This is then transferred to E1’s active-site cysteine residue in concert with the adenylylation of a second ubiquitin.[34] This adenylylated ubiquitin is then transferred to a cysteine of a second enzyme, ubiquitin-conjugating enzyme (E2).
In the last step, a member of a highly diverse class of enzymes known as ubiquitin ligases (E3) recognizes the specific protein to be ubiquitinated and catalyzes the transfer of ubiquitin from E2 to this target protein. A target protein must be labeled with at least four ubiquitin monomers (in the form of a polyubiquitin chain) before it is recognized by the proteasome lid.[35]
It is therefore the E3 that confers substrate specificity to this system.[36] The number of E1, E2, and E3 proteins expressed depends on the organism and cell type, but there are many different E3 enzymes present in humans, indicating that there is a huge number of targets for the ubiquitin proteasome system.
It seems to me that this system could not arise by small subsequent steps, and shows all signs of being intelligently designed.
https://reasonandscience.catsboard.com/t1851-proteasome-garbage-grinders
Proteasome:
1. The disposal of protein “trash” in the cell is the job of a complex machine called the proteasome. What could be more low than trash collection? Here also, sophisticated mechanisms work together.
2. PhysOrg described a new finding that shows that “two different mechanisms are required to determine which targets to destroy.” The “recognition tag” and “initiator tag.”
3. Both mechanisms have to be aligned properly to enter the machine’s disposal barrel. “The proteasome can recognize different plugs1, but each one has to have the correct specific arrangement of prongs1,” said a researcher at Northwestern University.
4. This is another example of interdependent irreducible complex systems. One can’t argue for evolution; that first only one system existed.
5. The work of a designer is again obvious and all men call him God.
6. God most probably exists.
https://www.youtube.com/watch?v=hvNJ3yWZQbE&t=90s
http://iaincarstairs.wordpress.com/2013/03/25/as-smart-as-molecules/

Once proteins are formed in the cell, there are circumstances in which they need to be destroyed, for example, if incorrectly formed or of no further use. The cells have their own aggressive TSA agents ( Transportation Security Administration ) who spot and stamp such proteins with a ubiquitin molecule which alerts other transport systems that the protein needs to be scrapped.
http://www.nature.com/ncb/journal/v6/n11/full/ncb1104-1011.html
Selective protein degradation by the ubiquitin–proteasome pathway is a fundamental regulatory strategy that fulfils essential roles in various cellular processes.
http://www.nature.com/ncb/journal/v7/n8/full/ncb0805-725.html
Following the discovery of ubiquitin in the late 1970s and the delineation of the enzymology of the pathway, its regulatory potential for orchestrating complex cellular programmes was initially severely underestimated.
How did the first cell " know " that a recycle mechanism for damaged proteins would be useful ? How did it either evolve the garbage grinder, or how could it possibly have arisen by chance ( since it had to be present probably as a essential mechanism right in the beginning of the first cell ? )
The proteosome : This shredder has a twist-open lid which is activated by the unique combination of three destruction tags added to the errant protein.
https://www.youtube.com/watch?v=hvNJ3yWZQbE
http://en.wikipedia.org/wiki/Proteasome#cite_note-Haas-34

Proteasomes are protein complexes inside all eukaryotes and archaea, and in some bacteria. In eukaryotes, they are located in the nucleus and the cytoplasm.The main function of the proteasome is to degrade unneeded or damaged proteins by proteolysis, a chemical reaction that breaks peptide bonds
Enzymes that carry out such reactions are called
proteases.
http://en.wikipedia.org/wiki/Protease
A protease (also termed peptidase or proteinase) is any enzyme that performs proteolysis, that is, begins protein catabolism by hydrolysis of the peptide bonds that link amino acids together in the polypeptide chain forming the protein.
Proteasomes are part of a major mechanism by which cells regulate the concentration of particular proteins and degrade misfolded proteins. The degradation process yields peptides of about seven to eight amino acids long, which can then be further degraded into shorter amino acid sequences and used in synthesizing new proteins. Proteins are tagged for degradation with a small protein called ubiquitin.
How did ubiquitin arise ? What function would it have, if not fully inserted in the whole mechanism to do the job ?
The tagging reaction is catalyzed by enzymes called ubiquitin ligases
http://en.wikipedia.org/wiki/Ubiquitin_ligase
A ubiquitin ligase (also called an E3 ubiquitin ligase) is a ligase enzyme that combines with a ubiquitin-containing E2 ubiquitin-conjugating enzyme, recognizes the target protein that is to be ubiquinated, and causes the attachment of ubiquitin to a lysine on the target protein via an isopeptide bond. An E3 ubiquitin ligase targets specific protein substrates for degradation by the proteasome. In general, the ubiquitin ligase is involved in poly-ubiquitination: a second ubiquitin is attached to the first, a third is attached to the second, and so forth. Poly-ubiquitination marks proteins for degradation by the proteasome.
Once a protein is tagged with a single ubiquitin molecule, this is a signal to other ligases to attach additional ubiquitin molecules. The result is a polyubiquitin chain that is bound by the proteasome, allowing it to degrade the tagged protein.[2]
In structure, the proteasome is a cylindrical complex containing a "core" of four stacked rings forming a central pore.
How did this structure arise ? what survival advantage would there be for a unfinished structure ?
Each ring is composed of seven individual proteins. The inner two rings are made of seven β subunits that contain three to seven protease active sites. These sites are located on the interior surface of the rings, so that the target protein must enter the central pore before it is degraded. The outer two rings each contain seven α subunits whose function is to maintain a "gate" through which proteins enter the barrel.
How did the barrel shape arise ?
These α subunits are controlled by binding to "cap" structures or regulatory particles that recognize polyubiquitin tags attached to protein substrates and initiate the degradation process. The overall system of ubiquitination and proteasomal degradation is known as the ubiquitin-proteasome system.
The proteasomal degradation pathway is essential for many cellular processes, including the cell cycle, the regulation of gene expression, and responses to oxidative stress. The importance of proteolytic degradation inside cells and the role of ubiquitin in proteolytic pathways was acknowledged in the award of the 2004 Nobel Prize in Chemistry to Aaron Ciechanover, Avram Hershko and Irwin Rose.[3]
In the first step, a ubiquitin-activating enzyme (known as E1) hydrolyzes ATP and adenylylates a ubiquitin molecule.
http://en.wikipedia.org/wiki/Ubiquitin-activating_enzyme
Ubiquitin-activating enzymes, also known as E1 enzymes, catalyze the first step in the ubiquitination reaction, which (among other things) can target a protein for degradation via a proteasome. This covalent attachment of ubiquitin or ubiquitin-like proteins to targeted proteins is a major mechanism for regulating protein function in eukaryotic organisms.[2] Many processes such as cell division, immune responses and embryonic development are also regulated by post-transcriptional modification by ubiquitin and ubiquitin-like proteins.
This is then transferred to E1’s active-site cysteine residue in concert with the adenylylation of a second ubiquitin.[34] This adenylylated ubiquitin is then transferred to a cysteine of a second enzyme, ubiquitin-conjugating enzyme (E2).
In the last step, a member of a highly diverse class of enzymes known as ubiquitin ligases (E3) recognizes the specific protein to be ubiquitinated and catalyzes the transfer of ubiquitin from E2 to this target protein. A target protein must be labeled with at least four ubiquitin monomers (in the form of a polyubiquitin chain) before it is recognized by the proteasome lid.[35]
It is therefore the E3 that confers substrate specificity to this system.[36] The number of E1, E2, and E3 proteins expressed depends on the organism and cell type, but there are many different E3 enzymes present in humans, indicating that there is a huge number of targets for the ubiquitin proteasome system.
It seems to me that this system could not arise by small subsequent steps, and shows all signs of being intelligently designed.
Last edited by Otangelo on Sat Jun 18, 2022 1:00 pm; edited 2 times in total




