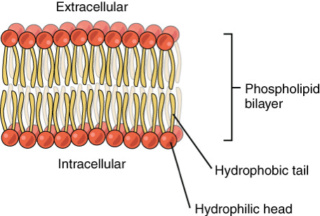
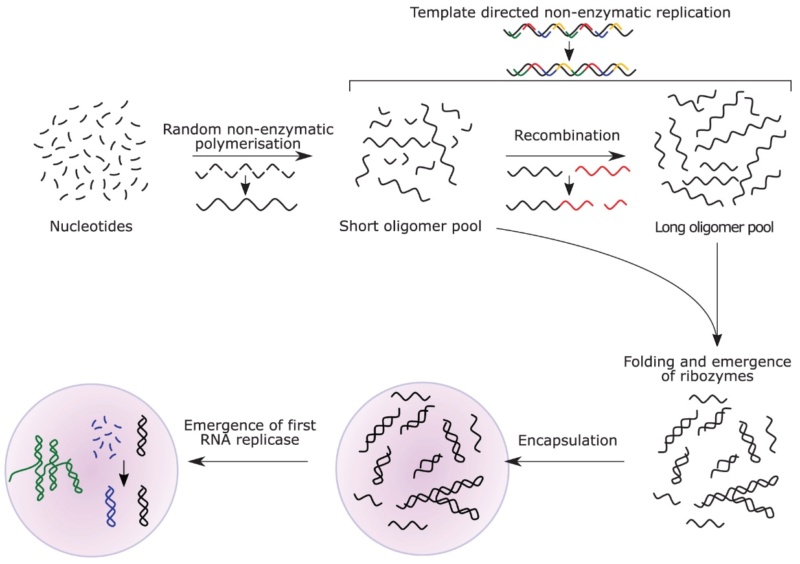
The Cell factory maker, Paley's watchmaker argument 2.0
https://www.amazon.com/Origin-Virus-World-Intelligent-Designer-ebook/dp/B0BJ9RYMD6?ref_=ast_sto_dp

https://reasonandscience.catsboard.com/t2809-on-the-origin-of-life-by-the-means-of-an-intelligent-designer
Index Page: On the origin of Cell factories by the means of an intelligent designer
Introduction
This book is about one of the deepest unsolved mysteries: The immense difficult puzzle of the origin of life. After at least 70 years of scientific inquiries and investigation, after Watson and Crick discovered the DNA molecule, and Miller & Urey performed their chemical experiment in 1953, which started the modern era of investigation of the origin of life, billions of dollars were spent, and millions of manhours invested to solve life's origin, it did not bring clear answers of the trajectory from non-life to life by chemical evolutionary means.
Kepa Ruiz-Mirazo (2014): We are still far from understanding which principles governed the transition from inanimate to animate matter. 15
Erik D Andrulis: (2011) Life is an inordinately complex unsolved puzzle. Despite significant theoretical progress, experimental anomalies, paradoxes, and enigmas have revealed paradigmatic limitations. 16
Yasuji Sawada (2020): The structures of biological organisms are miraculously complex and the functions are extremely multi-fold far beyond imagination. Even the global features such as metabolism, self-replication, evolution or common cell structures make us wonder how they were created in the history of nature. 16
Tomislav Stolar (2019): The emergence of life is among the most complex and intriguing questions and the majority of the scientific community agrees that we are far from being able to provide an answer to it 15
Finding a compelling explanation of how life started matters deeply to us since it is one of the relevant questions that can give us a hint in regards to the quest of which worldview is true: Naturalism or theism. Was a supernatural entity, an intelligent designer involved? Creation has been brand-marked by many as a myth. As fairy tale stories of ancient sheep herders, of uneducated tribesmen that lack the credentials compared to scientists of current days, which work actively in the field, and do the relevant scientific experiments, who can give a hint to the truth. Rather than revelation, science operates on experimentation. Is the hypothesis of Intelligent Design, not the case-adequate answer and explanation, after all?
Prof. Dr. Ruth E. Blake (2020): Investigating the origin of life requires an integrated and multidisciplinary approach including but not limited to biochemistry, microbiology, biophysics, geology, astrobiology, mathematical modeling, and astronomy. 14
Inquiry of the origin of life extends to several scientific disciplines: the research fields of synthetic organic chemists, biologists, biochemists, theoretical and evolutionary biologists, theoretical chemists, physical chemists, physicists, and earth and planetary scientists. Answering the question of the origin of life is maybe THE most challenging of all scientific open questions. Life is baffling. Cells are astonishingly complex. LIfe is staggeringly breathtakingly complex, fare beyond what any human technology has been able to come up with. The difference between life and non-life is mind-boggling. Cells are the most complex miniature chemical factories in the universe. They build their own building blocks, make their own energy, and host all the information to make all the awe-inspiringly complex machines, production lines, compartments, and information storage devices, and the envelope to host all the technology to make all this. Materials, energy, and information are joined to make a self-replicating factory. One of the three alone, on its own, achieves nothing. It has to be all three at the same time joined together as a team to climb mount insurmountable. Irreducible complex integrated, and interdependent factories are constructed by actualizing and instantiating instructional complex information that directs the cell making. Codified digital information directs and instantiates the equivalent analog device. But what came before the data? What instantiated transcription, and the code of translation? Where did all this come from?
Where did life come from? What is the origin of these masterfully crafted high-tech molecular machines, turbines, computers, and production lines? Science has, and still is facing monumental challenges in answering these questions. The author will lead in this book on a journey and hopes to lead the reader through its demonstration to the conclusion that intelligent design is very clear, and with no doubt, the best explanation. The evidence against naturalistic origins does not come from the outside critics like creationists, but from the inside out, from working scientists in the field. Peer-reviewed scientific papers demonstrate on their own that natural, nonguided, non-intelligent causes do not sufficiently, but inadequately explain the origin of life, constantly attempting to resort to natural selection and evolution, a mechanism that does not belong to the origin of life, and how evidence in special on a molecular level demonstrates overwhelmingly the signature and footprints of an intelligent, awe-inspiring, super-powerful designer. A mathematician, a physicist, a chemist, and a biologist, an engineer of the highest, finest, smartest, unfathomable order. One that created everything physical, instantiating and actualizing eternal energy, the laws that govern the universe, and creating life, complex and sophisticated, organized, integrated, and interdependent and complex from the start. Elegantly instantiating the most energy-efficient solutions, not leaving anything to chance, but designing and arranging every atom and placing it where it can perform its function, and contributing to the operation of the system as a whole. This book will unravel how many scientists ( mostly against their bias ) have to admit to having no compelling answers in regard to the Origin of Life (OoL). In the case of the origin of life, absence of evidence ( that unguided random events could in principle have selected and instantiated the first self-replicating, living cells ) is evidence of absence. The conclusion that an intelligent designer must have been involved in creating life, is not an argument from ignorance, but an inference to the best explanation based on all scientific information that we have at our disposition today. The facts that lead to the cause, the evidence is not provided by proponents of creationism and Intelligent Design but comes directly from the scientific, peer-reviewed literature. The authors in their smacking grand majority do hold to the inference that natural, and not supernatural causes explain all natural phenomena in question, but this is an inference that stands on philosophical, not scientific ground. Rather than actually finding explanations based on naturalistic causes, the gap is becoming wider and wider - the more science is advancing, and digging deeper into the intricacies, new levels of complexity are unraveled. In 1871, Nature magazine published an article about Ernst Haeckel's views on abiogenesis, describing Biological cells as essentially and nothing more than structureless bits of protoplasm without nuclei ( protoplasm = an albuminoid, nitrogenous Carbon compound ) as primitive slime - and their other vital properties can therefore simply and entirely brought about by the entirely by the peculiar and complex manner in which carbon under certain conditions can combine with the other elements. 1 This book aims, aside from other books already published on the subject, to advance the subject further, and bring less known facts to light, that illustrate the supreme sophistication of cells, and how Paley's watchmaker argument can be expanded to a new version, which we call: The factory maker argument.
Recent biochemistry books, like Bruce Alberts's classic: Molecular Biology Of The Cell 6 opened up an entirely new world of miniaturized technology on a microscale permitting to learn about the intricacies and complexity on a microscopic level, the inner workings inside of living cells, permitting a journey into a microworld, that is bewildering, full of unexpected, awe-inspiring high-tech devices, and incredible worlds that scientists, a half century back, never thought to encounter. There are a wealth of mindboggling aspects of life, the superb engineering & architecture of cells, and their precise exquisite operations, that are not common knowledge. They point to a super-intellect, and engineer of unimaginable intelligence and power. One of our awe-inspiring discoveries over the years was the fact that cells are not analogous to factories, but an entire city, park, or quartier of interlinked chemical high-tech factories in a literal sense with superb capabilities, efficiency, and adaptive design. Dozens and dozens of scientific papers describe cells as literal minuscule autonomous self-replicating factories, that build and operate based on information, and sophisticated interconnected signaling networks. There are two types of information. Descriptive, and prescriptive. Descriptive information is when someone encounters an artifact, and describes it. Blueprints, floorplans, architectural drawings, and a recipe to make a cake, all contain information on " know-how". It is instructional assembly information. It instructs, dictates, directs, orients, and tells the reader/receiver how to construct, how assemble, and how to make or how operate things. Interestingly, that is precisely the kind of information that we encounter in the genome and epigenetic information storage mechanisms. Some have described the genome as a blueprint, and others compared it to a library. A scrambled and seemingly disorganized collection of genes, analogous to books. Most of it is so-called Junk-DNA, while just a tiny part, a few percent, encodes proteins, the working horses of the cell. Time has more and more unraveled, that this so-called Junk DNA is actually not non-functional junk, a remnant from a long evolutionary past, but it has a function. It regulates gene expression, amongst other tasks.

In the last 150 years, since Haeckel's ink, the understanding on a molecular level of how cells operate and function has gone through a remarkable development. The advance of scientific investigation has come to realize, what was well expressed in an article in 2014:
The upward trend in the literature reveals that as technology allows scientists to investigate smaller and smaller cellular structures with increasing accuracy, they are discovering a level of molecular complexity that is far beyond what earlier generations had predicted. Molecular biologists often focus years of their research on individual molecules or pathways. 2
What science has come to discover, is more and more new levels of complexity down to the precise complex specified order, arrangement, and fine-tuning of the most basic molecules and building blocks used in life down to atoms.
Bharath Ramsundar gave some data points in a remarkable article. He wrote in in 2016:
One neuron contains roughly 175 trillion ( precisely ordered) atoms per cell. Assuming that the brain contains 100 billion neurons, the total brain contains roughly 2 * 10^25 atoms. ( That's more than the number of stars in the universe ) Both estimates are likely low. 3
Another example we find in David Goodsell's excellent book: Our Molecular Nature, page 26:
Dozens of enzymes are needed to make the DNA bases cytosine and thymine from their component atoms. The first step is a "condensation" reaction, connecting two short molecules to form one longer chain, performed by aspartate carbamoyltransferase. Other enzymes then connect the ends of this chain to form the six-sided ring of nucleotide bases, and half a dozen others shuffle atoms around to form each of the bases. In bacteria, the first enzyme in the sequence, aspartate carbamoyltransferase, controls the entire pathway. (In human cells, the regulation is more complex, involving the interaction of several of the enzymes in the pathway.) Bacterial aspartate carbamoyltransferase determines when thymine and cytosine will be made, through a battle of opposing forces. It is an allosteric enzyme, referring to its remarkable changes in shape (the term is derived from the Greek for "other shape"). The enzyme is composed of six large catalytic subunits and six smaller regulatory subunits. Take just a moment to ponder the immensity of this enzyme. The entire complex is composed of over 40,000 atoms, each of which plays a vital role. The handful of atoms that actually perform the chemical reaction are the central players. But they are not the only important atoms within the enzyme--every atom plays a supporting pan. The atoms lining the surfaces between subunits are chosen to complement one another exactly, to orchestrate the shifting regulatory motions. The atoms covering the surface are carefully picked to interact optimally with water, ensuring that the enzyme doesn't form a pasty aggregate, but remains an individual, floating factory. And the thousands of interior atoms are chosen to fit like a jigsaw puzzle, interlocking into a sturdy framework. Aspartate carbamoyltransferase is fully as complex as any fine automobile in our familiar world. And, just as manufacturers invest a great deal of research and time into the design of an automobile, enzymes like aspartate carbamoyltransferase have been finely tuned and now, Goodsell adds just five words at the end of the sentence - over the course of evolution.. 4
After describing like an epic - how this enzyme is masterfully crafted down to the atomic scale, resorting to the analogy of the manufacturing of an automobile, he concludes that this life-essential enzyme was finely tuned by a process that does not operate based on intelligence - namely evolution. You did read that right. Worse than that: Aspartate Carbamoyltransferase is part of the biosynthesis of pyrimidines - the nucleobases that make up RNA and DNA, life-essential molecules that had to be made prior to when life started, and as such, evolution as the mechanism to synthesize them cannot be invoked. There is a minimal number of proteins and enzymes that are required to start life, and the synthesis of RNA and DNA is life essential, and as such, the origin of Aspartate Carbamoyltransferase is a question that belongs to the origin of life research.
There is more: Remarkably, not only this exquisitely engineered molecular machine that makes nucleobases is finely and precisely adjusted to operate and perform its job on an atomic scale: Its product is finely tuned as well to perform a life-essential function: Forming Watson-Crick base-pairing, giving rise to the famous DNA ladder, the information storage mechanism of life.
John D. Barrow reports in his book: FITNESS OF THE COSMOS FOR LIFE, Biochemistry and Fine-Tuning, on page 154:
Today, it is particularly striking to many scientists that cosmic constants, physical laws, biochemical pathways, and terrestrial conditions are just right for the emergence and flourishing of life. It now seems that only a very restricted set of physical conditions operative at several major junctures of emergence could have opened the gateways to life. Fine-tuning in biochemistry is represented by the strength of the chemical bonds that makes the universal genetic code possible. Neither transcription nor translation of the messages encoded in RNA and DNA would be possible if the strength of the bonds had different values. Hence, life, as we understand it today, would not have arisen. As it happens, the average bond energy of a carbon-oxygen double bond is about 30 kcal per mol higher than that of a carbon-carbon or carbon-nitrogen double bond, a difference that reflects the fact that ketones normally exist as ketones and not as their enol-tautomers. If (in the sense of a “counterfactual variation”) the difference between the average bond energy of a carbon-oxygen double bond and that of a carbon-carbon and carbon-nitrogen double bond were smaller by a few kcal per mol, then the nucleobases guanine, cytosine, and thymine would exist as “enols” and not as “ketones,” and Watson–Crick base-pairing would not exist – nor would the kind of life we know. It looks as though this is providing a glimpse of what might appear (to those inclined) as biochemical fine-tuning of life. 5
The machinery that makes RNA and DNA, aspartate carbamoyltransferase, is finely tuned on an atomic level - depending on the information stored in DNA to be made. The nucleobases that make up DNA, also finely tuned in their atomic arrangement to permit the right forces to make Watson-Crick base-pairing possible, and so, the stable life-essential information storage mechanism depends on the finely tuned aspartate carbamoyltransferase machine to be synthesized - that creates a Catch22 situation. If everything depends on everything to be made, how did it all start?
To me, fine-tuning, interdependence, irreducible complexity, and complex specified order on an atomic scale is awe-inspiring evidence of creation. Maybe the skeptical reader will react with resistance to such evidence and will attempt to resort to naturalistic, non-engineered principles acting in nature. The author will provide many other examples that will make it very difficult - if not impossible - to keep asserting that non-intelligent mechanisms are a compelling explanation. For some, there is never enough evidence to conclude God. But others might come to realize that the teleological argument today is solidly grounded in scientific evidence, and as such, it is reasonable to be an advocate of intelligent design.
Chapter 1
The aim of this book is not to point to a specific divinity, but to an intelligent designer as the best explanation of the origins of life, and biodiversity. Identifying the creator or which religion is true is not the scope of this book. The author is relying on peer-reviewed scientific papers to make his case. Providing these sources is used as the basis of the conclusive arguments. A common criticism is that every scientific problem, where God was inserted in a gap of knowledge, has been replaced with a scientific one. The more science advances, the more it solves open questions, and religion is receding and hiding into an ever-smaller getting pocket of ignorance. "In the past, we didn't know how lightning worked, and Zeus was invoked. Today we know better." We hear that frequently. The conflict has been portrayed as between Science and religion. Science is based on reason and the scientific method, which gives us empirical results. Religion is based on blind faith, and left to the "religtards". Reason-based thinking is supposedly based on apistevism, with no faith required ( Not considering, that whatever position one takes, naturalism, or theism, is a position one has to take on faith, since nobody has access to the realm beyond our physical universe. Metaphysics and the realm of fundamental ontology are beyond scientific investigation. Religion vs science is a false dichotomy. It's not rare that when I ask an atheist that denies a creator, what he would like to replace God with, the answer is often: Science...... not recognizing the logical fallacy that science is not a causal mechanism or agency, but a tool to explore reality. Kelly James Clark writes in his book: Religion and the Sciences of Origins: Historical and Contemporary Discussions:
The deepest intellectual battle is not between science and religion (which, as we have seen, can operate with a great deal of accord), but between naturalism and theism—two broad philosophical (or metaphysical) ways of looking at the world. Neither view is a scientific view; neither view is based on or inferable from empirical data. Metaphysics, like numbers and the laws of logic, lies outside the realm of human sense experience. So the issue of naturalism versus theism must be decided on philosophical grounds. Metaphysical naturalism is the view that nothing exists but matter/energy in space-time. Naturalism denies the existence of anything beyond nature. The naturalist rejects God, and also such spooky entities as souls, angels, and demons. Metaphysical naturalism entails that there is no ultimate purpose or design in nature because there is no Purposer or Designer. On the other hand, theism is the view that the universe is created by and owes its sustained existence to a Supreme Being that exists outside the universe. These two views, by definition, contradict each other. 6
The proposition that no creator is required to explain our existence, is what unites all nonbelievers, weak, strong atheists, agnostics, skeptics, and nihilists. When pressed hard, how that makes sense, the common cop-out is: We don't know what replaces God. Science is working on it.
God of the gaps
Isaac Newton and Laplace did provide a classic example of a God-of-the-gaps argument. Newton's equations were a great tool to predict and explain the motions of planets. Since there are several gravitational interactions between them, Newton suspected that gravity would interrupt their trajectories, and God would intervene from time to time to solve the problem and bring them back on track. A legend tells us Laplace who was a french mathematician, was brought to Napoleon who asked him about the absence of God in his theory: “M. Laplace, they tell me you have written this large book on the system of the universe, and have never even mentioned its Creator.” To this, Laplace famously replied, “I had no need of that hypothesis.” Newton used his lack of knowledge to insert God in the gap. It becomes problematic when later, the gap is filled with a scientific explanation. Design advocates are frequently accused of using this fallacy. The argument that God is a gap filler is really boring, a beaten horse ad nauseam. Its invoked in almost every theist-atheist debate when atheists are unable to successfully refute a theist claim. No, God is NOT a gap filler. God can be a logical inference based on the evidence observed and at hand in the natural world. If a theist would say, ''We don't know what caused 'x', therefore, God.'', it would be indeed a God of the gaps fallacy. What, however, today, often can be said, is: ''Based on current knowledge, an intelligent creative agency is a better explanation than materialistic naturalism." If one is not arguing from ignorance, but rather reasoning from the available evidence to the best explanation, is it not rather ludicrous to accuse ID proponents of launching a 'god of the gaps argument'?
Paul Davies comments in The Goldilocks enigma, on page 206
The weak point in the “gaps” argument of the Intelligent Design movement is that there is no reason why biologists should immediately have all the answers anyway. Just because something can’t be explained in detail at this particular time doesn’t mean that it has no natural explanation: it’s just that we don’t know what it is yet. Life is very complicated, and unraveling the minutiae of the evolutionary story in detail is an immense undertaking. Actually, in some cases we may never know the full story. Because evolution is a process that operates over billions of years, it is entirely likely that the records of many designlike features have been completely erased. But that is no excuse for invoking magic to fill in the gaps. So could it be that life’s murky beginning is one of those “irreducible” gaps in which the actions of an intelligent designer might lie? I don’t think so. Let me repeat my warning. Just because we can’t explain how life began doesn’t make it a miracle. Nor does it mean that we will never be able to explain it—just that it’s a hard and complicated problem about an event that happened a long time ago and left no known trace. But I for one am confident that we will figure out how it happened in the not-too-distant future. 7
Isn't that interesting? While Davies accuses ID proponents of using gap arguments, he does precisely that, by inserting naturalism into gaps of knowledge. We don't know (yet), therefore natural processes must have done it. Why can someone not start with the presupposition that an eternal, powerful, intelligent conscious creator must be at the bottom of reality, and use that as a starting point to investigate if the God hypothesis withstands scrutiny?
Limited causal alternatives do not justify to claim of " not knowing "
Hosea 4:6 says: People are destroyed for lack of knowledge. People, often, either because of confirmation bias or bad will, blindly believe what others say, without scrutinizing on their own, if what they read and are informed about, is true, or made-up, unwarranted, and based on superficial, and at the end, false, misleading information. Not only that. When questioned, they argue based on that badly researched information and expect others to take them seriously, and regard them as knowledgeable. This is in particular frustrating when the interlocutor actually HAS investigated the issue in demand in a duly, serious fashion, knows the issue at hand, but encounters deaf ears. Scientists HATE saying "we don't know. " They prefer to shut their face until they do know. Much less base their entire worldview on being "comfortable of not knowing". Confessing of not knowing, when there are good reasons to confess ignorance, is justifiable. But claiming not knowing something, despite the evident facts easy at hand and having the ability to come to informed well-founded conclusions based on sound reasoning, and through known facts and evidence, is not only willful ignorance but plain foolishness. Especially, when the issues in the discussion are related to origins and worldviews, which ground how we see the world, past present, future, morals, etc. If there were hundreds of possible statements, then claiming not knowing which makes most sense could be justified. In the quest of God, there are just two possible explanations.
A God, or no God. That's the question
Either there is a God-creator and causal agency of the universe, or not. God either exists or he doesn’t. These are the only two possible explanations. The law of excluded middle is given the name of law for a reason. It's called so when we say something is either A or it is not A there's no middle there; no third option; it is one of the fundamental laws of logic. It's a dichotomy: it's either God or not God. When we reduce the "noise", we come down to what the distinction is between the two big competing worldviews. Either nature is the product of pointless stupidity of no existential value or the display of God's sublime grandeur and intellect. How we answer the God Question has significant implications for how we understand ourselves, our relation to others, and our place in the universe. Remarkably, however, many people [ in special in the West, like in Europe, for example ] don’t give this question nearly the attention it deserves; they live as though it doesn’t really matter to everyday life. Either our worldview is based on believing in naturalism & materialism, which means that the physical world had no causal agency, or our worldview is based on deism, theism & or creationism. Some posit a pantheistic principle (impersonal spirit) but in the author's view, spirits are by definition personal. That is the dichotomy that simplifies our investigation a lot: Or an intelligent designer, a powerful creator, a conscious being with will, foresight, aims, and goals exists, or not.
What's the Mechanism of Intelligent Design?
We don't know how exactly a mind might act in the world to cause change. Your mind, mediated by your brain, sends signals to your arm, hand, and fingers, and writes a text through the keyboard of the computer I sit here typing. I cannot explain to you how exactly this process functions, but we know, it happens. Consciousness can interact with the physical world and cause change. But how exactly that happens, we don't know. Why then should we expect to know how God created the universe? The hypothesis of intelligent design proposes an intelligent mental cause as the origin of the physical world. Nothing else.
Is the "God concept" illogical?
The author has seen several atheists claiming that they were never presented with a God concept that was possible, logical, plausible, and not contradictory - and therefore, impossible. The author's description of God for this discussion is: God is a conscious mind. Is spaceless, timeless, immaterial, powerful, intelligent, and personal, which brought space, time, and matter into being. Consciousness englobes the mind, "qualia", intellectual activity, calculating, thinking, forming abstract ideas, imagination, introspection, cognition, memories, awareness, experiencing, intentions, free volition, free creation, invention, and generation of information. It classifies, recognizes, and judges behavior, good and evil. It is aware of beauty and feels sensations and emotions. There is nothing contradictory in this hypothesis, and it is therefore on the table as a possible option.
Did God create Ex-nihilo?
God could be eternally in the disposition of infinite potential energy and disposes of it whenever it fits him to use it. He can both be an eternal mind of infinite knowledge, wisdom, and intelligence, that creats minds similar to his own mind, but with less, and limited intelligence, but also physical worlds/universes through his eternal power. Power comes from Latin, and means, potere. Potere means able of doing. Able to provoke change. God is spirit, but has eternal power at his disposal to actualize it in various forms. So basically, he is not creating the physical world from nothing, but from a potential at his disposal, which can manifest when he decides so, precisely in the way that he wills and decides to do. So anything external to him is instantiated, uphold, and secured by his eternal power.
The potential of energy was/is with God and when God created our universe, in the first instant, he focussed and concentrated enormous power or energy that is at his disposition, into a single point, a singularity, which triggered the creation and stretching out our universe. The temperatures, densities, and energies of the Universe would be arbitrary, unimaginably large, and would coincide with the birth of time, matter, and space itself, and God subdued and ordained the energy to start obeying the laws of physics that he instantiated at the same time. We know that matter/energy are interchangeable. There had to be a connection between God and the Universe. God did not only create the universe, but sustains it permanently through his power, and ordains the laws of physics to impose and secure that the universe works orderly, with stability, and in a predictable manner.
Intelligence vs no intelligence
How can we recognize the signature of (past) intelligent actions? Contrasting and comparing "intended" versus "accidental" arrangements leads us to the notion of design. We have an extensive experience-based understanding of the kinds of things that intelligent minds design to instantiate devices for specific purposes and functions. We also know by experience the range of stochastic accidental natural phenomena and causes, and what they can instantiate. Opponents commonly object and argue that intelligent design does not make predictions. But since we have already background knowledge of what intelligent minds can do, we can attribute similar effects, to similar causes. A physical system is composed of a specific arrangement of matter and parts: a machine, a car, a clock. When we describe it and quantify its size, structure, and motions, and annotate the materials used, that description contains information. When we arrange and distribute materials in a certain way for intended means, we can produce things for specific aims and call them design. The question is, therefore, when we see things in nature that appear to have specific functions and give the impression of being designed, ARE they indeed the product of intentional design? Science has unraveled structures of mind-boggling organizational intricacies at the molecular level that leave us in awe, so sophisticated that our most advanced technology seems kindergarten by comparison. A proponent of naturalism has to posit that physical and chemical things emerged by chance and/or physical necessity and biological systems by mindless evolutionary pressures. These organic structures like living cells present us with a degree of complexity that science has failed to explain stochastically by unguided means. No scientific experiment has been able to come even close to synthesizing the basic building blocks of life and reproducing a self-replicating Cell in the Laboratory through self-assembly and autonomous organization. The total lack of any kind of experimental evidence leading to the re-creation of life; not to mention the spontaneous emergence of life… is the most humiliating embarrassment to the proponents of naturalism and the whole so-called “scientific establishment” around it… because it undermines the worldview of who wants naturalism to be true. Everything we know tells us that machines, production lines, computers, energy generating turbines, transistors, and interlinked factories are structures designed by an intelligence. The cooperation and interdependent action of proteins and co-factors in cells is stupendous and depends on very specific controlled and arranged mechanisms, precise allosteric binding sites, and finely-tuned forces. Accidents do not design machines. Intellect does. Intelligence leaves behind a characteristic signature. The action or signature of an intelligent designer can be detected when we see :
1. Implementing things conveying regular behavior, order, stability, and predictability. The imposition of mathematical rules, laws, principles, and physical constants.
2. Something purposefully and intentionally developed and made to accomplish a specific goal(s). That includes specifically the generation and making of building blocks, energy, and the instantiation of instructional blueprints, floorplans, and complex specified information. If an arrangement of parts is a) perceptible by a reasonable person as having a purpose and/or a function, and b) can be used for the perceived purpose then its purpose was designed by an intelligent mind.
3. Repeating a variety of complex actions with precision based on methods that obey instructions, governed by rules.
4. An instructional complex blueprint (bauplan) or protocol to make objects ( machines, factories, houses, cars, etc.) that are irreducible complex, integrated, and an interdependent system or artifact composed of several interlocked, well-matched hierarchically arranged systems of parts contributing to a higher end of a complex system that would be useful only in the completion of that much larger system. The individual subsystems and parts are neither self-sufficient, and their origin cannot be explained individually, since, by themselves, they would be useless. The cause must be intelligent and with foresight, because the unity transcends every part, and thus must have been conceived as an idea, because, by definition, only an idea can hold together elements without destroying or fusing their distinctness. An idea cannot exist without an intelligent creator, so there must be an intelligent mind.
5. Artifacts which use might be employed in different systems ( a wheel is used in cars and airplanes )
6. Things that are precisely adjusted and finely tuned to perform precise functions and purposes.
7. Arrangement of materials and elements into details, colors, and forms to produce an object or work of art able to transmit the sense of beauty, and elegance, that pleases the aesthetic senses, especially the sight. “I declare this world is so beautiful that I can hardly believe it exists.”
8. Establishing a language, code, communication, and information transmission system, that is 1. A language, 2. the information (message) produced upon that language, the 3 .information storage mechanism ( a hard disk, paper, etc.), 4. an information transmission system, that is: encoding - sending and decoding) and eventually fifth, sixth, and seventh ( not essential): translation, conversion, and transduction.
9. Any scheme where instructional information governs, orchestrates, guides, and controls the performance of actions of constructing, creating, building, and operating. That includes operations and actions as adapting, choreographing, communicating, controlling product quality, coordinating, cutting, duplicating, elaborating strategies, engineering, error checking and detecting, and minimizing, expressing, fabricating, fine-tuning, foolproof checking, governing, guiding, implementing, information processing, interpreting, interconnecting, intermediating, instructing, logistic organizing, managing, monitoring, optimizing, orchestrating, organizing, positioning, monitoring and managing of quality, regulating, recruiting, recognizing, recycling, repairing, retrieving, shuttling, separating, self-destructing, selecting, signaling, stabilizing, storing, translating, transcribing, transmitting, transporting, waste managing.
10. Designed objects exhibit “constrained optimization.” The optimal or best-designed laptop computer is the one that is the best balance and compromises multiple competing factors.
Now lets see if we observe the signature of intelligent action in nature:
1. Paul Davies: The universe is governed by dependable, immutable, absolute, universal, mathematical laws of an unspecified origin.
Eugene Wigner: The mathematical underpinning of nature "is something bordering on the mysterious and there is no rational explanation for it.
Richard Feynman: Why nature is mathematical is a mystery...The fact that there are rules at all is a kind of miracle.
Albert Einstein: How can it be that mathematics, being, after all, a product of human thought which is independent of experience, is so admirably appropriate to the objects of reality?
Max Tegmark: Nature is clearly giving us hints that the universe is mathematical.
2. Basically, everything in biology is purposeful [ in contrast to the structure of a rock, for example], and has a specific function. Examples abound. Like co-factors and apo-proteins ( lock and key). Cells are interlocked irreducible factories where a myriad of proteins work together to self-sustain and perpetuate life. To replicate, reproduce, adapt, grow, remain organized, store, and use the information to control metabolism, homeostasis, development, and change. A lifeless Rock has no goal, has no specific shape or form for a specific function, but is random, and the forms of stones and mountains come in all chaotic shapes, sizes, and physicochemical arrangements, and there is no goal-oriented interaction between one rock and another, no interlocking mechanical interaction.
3. A variety of biological events are performed in a repetitive manner, described in biomechanics, obeying complex biochemical and biomechanical signals. These include, for example, cell migration, cell motility, traction force generation, protrusion forces, stress transmission, mechanosensing and mechanotransduction, mechanochemical coupling in biomolecular motors, synthesis, sorting, storage, and transport of biomolecules
4. In living cells, information is encoded through at least 30 genetic, and almost 30 epigenetic codes that form various sets of rules and languages. They are transmitted through a variety of means, that is the cell cilia as the center of communication, microRNA's influencing cell function, the nervous system, the system synaptic transmission, neuromuscular transmission, transmission b/w nerves & body cells, axons as wires, the transmission of electrical impulses by nerves between brain & receptor/target cells, vesicles, exosomes, platelets, hormones, biophotons, biomagnetism, cytokines and chemokines, elaborate communication channels related to the defense of microbe attacks, nuclei as modulators-amplifiers. These information transmission systems are essential for keeping all biological functions, that is organismal growth and development, metabolism, regulating nutrition demands, controlling reproduction, homeostasis, constructing biological architecture, complexity, form, controlling organismal adaptation, change, regeneration/repair, and promoting survival.
5. Some convergent systems are bat echolocation in bats, oilbirds, and dolphins, cephalopod eye structure, similar to the vertebrate eye, an extraordinary similarity of the visual systems of sand lance (fish) and chameleon (reptile). Both the chameleon and the sand lance move their eyes independent of one another in a jerky manner, rather than in concert. Chameleons share their ballistic tongues with salamanders and sand lace fish. There are a variety of organisms, unrelated to each other, that encounter nearly identical convergent biological systems. This commonness makes little sense in light of evolutionary theory. If evolution is indeed responsible for the diversity of life, one would expect convergence to be extremely rare.
6. The Universe is wired in such a way that life in it is possible. That includes the fine-tuning of the Laws of physics, the physical constants, the initial conditions of the universe, the Big Bang, the cosmological constant, the subatomic particles, atoms, the force of gravity, Carbon nucleosynthesis, the basis of all life on earth, the Milky Way Galaxy, the Solar System, the sun, the earth, the moon, water, the electromagnetic spectrum, biochemistry. Hundreds of fine-tuning parameters are known. Even in biology, we find fine-tuning, like nucleobase isomeric arrangement that permits Watson-Crick base-pairing, cellular signaling pathways, photosynthesis, etc.
7. I doubt someone would disagree with Ralph Waldo Emerson. Why should we expect beauty to emerge from randomness? If we are merely atoms in motion, the result of purely unguided processes, with no mind or thought behind us, then why should we expect to encounter beauty in the natural world, and the ability to recognize beauty, and distinguish it from ugliness? Beauty is a reasonable expectation if we are the product of design by a designer who appreciates beauty and the things that bring joy.
8. In the alphabet of the three-letter word found in cell biology are the organic bases, which are adenine (A), guanine (G), cytosine (C), and thymine (T). It is the triplet recipe of these bases that make up the ‘dictionary’ we call molecular biology genetic code. The code system enables the transmission of genetic information to be codified, which at the molecular level, is conveyed through genes. Pelagibacter ubique is one of the smallest self-replicating free-living cells, has a genome size of 1,3 million base pairs, and codes for about 1,300 proteins. The genetic information is sent through communication channels that permit encoding, sending, and decoding, done by over 25 extremely complex molecular machine systems, which do as well error checks and repair to maintain genetic stability, and minimizing replication, transcription, and translation errors, and permit organisms to pass accurately genetic information to their offspring, and survive.
9. Science has unraveled, that cells, strikingly, are cybernetic, ingeniously crafted cities full of interlinked factories. Cells contain information, which is stored in genes (books), and libraries (chromosomes). Cells have superb, fully automated information classification, storage, and retrieval programs ( gene regulatory networks ) that orchestrate strikingly precise and regulated gene expression. Cells also contain hardware - a masterful information-storage molecule ( DNA ) - and software, more efficient than millions of alternatives ( the genetic code ) - ingenious information encoding, transmission, and decoding machinery ( RNA polymerase, mRNA, the Ribosome ) - and highly robust signaling networks ( hormones and signaling pathways ) - awe-inspiring error check and repair systems of data ( for example mind-boggling Endonuclease III which error checks and repairs DNA through electric scanning ). Information systems, which prescribe, drive, direct, operate, and control interlinked compartmentalized self-replicating cell factory parks that perpetuate and thrive life. Large high-tech multimolecular robotlike machines ( proteins ) and factory assembly lines of striking complexity ( fatty acid synthase, non-ribosomal peptide synthase ) are interconnected into functional large metabolic networks. In order to be employed at the right place, once synthesized, each protein is tagged with an amino acid sequence, and clever molecular taxis ( motor proteins dynein, kinesin, transport vesicles ) load and transport them to the right destination on awe-inspiring molecular highways ( tubulins, actin filaments ). All this, of course, requires energy. Responsible for energy generation are high-efficiency power turbines ( ATP synthase )- superb power generating plants ( mitochondria ) and electric circuits ( highly intricate metabolic networks ). When something goes havoc, fantastic repair mechanisms are ready in place. There are protein folding error check and repair machines ( chaperones), and if molecules become non-functional, advanced recycling methods take care ( endocytic recycling ) - waste grinders and management ( Proteasome Garbage Grinders )
10. Translation by the ribosome is a compromise between the opposing constraints of accuracy and speed.
Genesis or Darwin?
In the west, the tradition of the Judeo-Christian God revealed in the Bible shaped the Worldview of many generations, and the greatest pioneers of science like Kepler, Galileo, Newton, Boyle, Maxwell, etc. were Christians, and firmly believed that a powerful creator instantiated the natural order. Butterfield puts it that way:
Until the end of the Middle Ages there was no distinction between theology and science. Knowledge was deduced from self-evident principles received from God, so science and theology were essentially the same fields. After the Middle Ages, the increasingly atheistic rejection of God by scientists led to the creation of materialist secular science in which scientists will continue to search for a natural explanation for a phenomenon based on the expectation that they will find one. 14
Naturalists hijack science by imposing philosophical naturalism
From 1860 - to 1880, Thomas Huxley and members of a group called the “X Club” effectively hijacked science into a vehicle to promote materialism (the philosophy that everything we see is purely the result of natural processes apart from the action of any kind of god and hence, science can only allow natural explanations). Huxley was a personal friend of Charles Darwin, who was more introverted and aggressively fought the battle for him. Wikipedia has an interesting article worth reading titled, “X Club.” It reveals a lot about the attitudes, beliefs, and goals of this group. 8
The fact that science papers do not point to God, does not mean that the evidence unraveled by science does not point to God. All it means is that the philosophical framework based on methodological naturalism that surrounds science since its introduction in the 19th century is a flawed framework, and should have been changed a long time ago when referencing historical science, which responds to questions of origins. Arbitrary a priori restrictions are the cause of bad science, where it is not permitted to lead the evidence wherever it is. The proponents of design make only the limited claim that an act of intelligence is detectable in the organization of living things, and using the very same methodology that materialists themselves use to identify an act of intelligence, design proponents have successfully demonstrated their evidence. In turn, their claim can be falsified with a single example of a dimensional semiotic system coming into existence without intelligence.
Sean Carroll writes:
Science should be interested in determining the truth, whatever that truth may be – natural, supernatural, or otherwise. The stance known as methodological naturalism, while deployed with the best of intentions by supporters of science, amounts to assuming part of the answer ahead of time. If finding truth is our goal, that is just about the biggest mistake we can make. 9
Scientific evidence is what we observe in nature. The understanding of it like microbiological systems and processes is the exercise and exploration of science. What we infer through the observation, especially when it comes to the origin of given phenomena in nature, is philosophy, and based on individual induction and abductive reasoning. What looks like a compelling explanation to somebody, can not be compelling to someone else, and eventually, I infer the exact contrary. In short, the imposition of methodological naturalism is plainly question-begging, and it is thus an error of method. No one can know with absolute certainty that the design hypothesis is false. It follows from the absence of absolute knowledge, that each person should be willing to accept at least the possibility that the design hypothesis is correct, however remote that possibility might seem to him. Once a person makes that concession, as every honest person must, the game is up.
In Genesis, all life forms were created, all creatures according to their kinds. It is stated as a fact. In 1837, Charles Darwin draw a simple tree in one of his notebooks, and above, he wrote: “I think.”
What we see right here, is the difference between stating something as a fact, and another as uncertain hypothetical imagination. While Genesis as the authoritative word of God makes absolute claims, man, in his limitless, can only speculate, infer, and express what he thinks to be true based on circumstantial evidence. Darwin incorporated the idea in: On the Origin of Species (1859), where he wrote:
The affinities of all the beings of the same class have sometimes been represented by a great tree. I believe this simile largely speaks the truth. 10
Since then, science, and biology textbooks have adopted Darwin's view of universal common descent, denying the veracity of the biblical Genesis account, and replaced it with the evolutionary narrative to explain the diversity of life.
Creationist objections have often been refuted by resorting to scientific consensus, claiming that evolution is a fact, disregarding that the term evolution, before using it, has to be defined. More than that. Intelligent design has been brand marked as pseudo-science and been rejected by the scientific community.11
In nature, life comes only from life. That has never been disproven. Therefore, that should in our view be the default position. In this book, we will show how eliminative induction refutes the origin and complexification of life by natural means, and how abductive reasoning to the best explanation leads to an intelligent designer as the best explanation of the origin and diversification of life. Origin of life research in over 70 years has led only to dead ends. Furthermore, every key branch of Darwin's tree of life is fraught with problems, starting with the root of the tree of life, to every major key transition. From the first to the last universal common ancestor, to the three domains of life, from unicellular to multicellular life. Wherever one looks, there are problems. Nothing in Biology Makes Sense Except in the Light of Evolution was Dobzhansky's famous dictum back in 1973. The author's view is that nothing in physics, chemistry, and biology makes sense except in the Light of intelligent design by a superintelligent powerful creator, that made everything for his own purposes, and his glory. Louis Pasteur, famously stated: "Little Science takes you away from God, but more of it takes you to him". The author agrees with Pasteur. For over 160 years, Darwin's Theory of Evolution has influenced and penetrated biological thinking and remains the dominant view of the history of life in academia, and in general. Despite its popularity, the Bible, which is in disagreement with Darwin's view, is still believed to be true by a large percentage of the population in the united states. The Genesis account states that God created the universe, and the world, in literally six days, and each of its living kind individually. Both accounts, which contradict each other, cannot be true. If one is true the other must be false. The dispute between them is an old one. Ultimately, each one of us has to find out individually, what makes the most sense. In this book, we use the approach of eliminative induction, and bayesian thinking, abductive reasoning, inference to the best explanation, to come to the conclusion that design tops naturalistic explanations like evolution when it comes to explaining the origin of life, and biodiversity.
Intelligent design wins using eliminative induction based on the fact that its competitors are false. Materialism explains basically nothing consistently in regards to origins but is based on unwarranted consensus and scientific materialism, a philosophical framework, that should never have been applied to historical sciences. Evidence should be permitted to lead wherever it is. Also, eventually, an intelligent agency is the best explanation of origins.
And it wins based on abductive reasoning, using inference to the best explanation, relying on positive evidence, on the fact that basically, all-natural phenomena demonstrate the imprints and signature of intelligent input and setup. We see an unfolding plan, a universe governed by laws, that follows mathematical principles, finely adjusted on all levels, from the Big Bang to stars, galaxies, the earth, to permit life, which is governed by instructional complex information stored in genes and epigenetically, encoding, transmitting and decoding information, used to build, control and maintain molecular machines ( proteins ) that are build based on integrated functional complex parts, which are literally nanorobots with internal communication systems, fully automated manufacturing production lines, transport carriers, turbines, transistors, computers, and factory parks, employed to give rise to a wide range, millions of species, of unimaginably complex multicellular organisms. This book will focus on how the cell, which is the smallest unit of life, provides the most fascinating and illustrative evidence of design in nature, and so, pointing to an intelligent designer.
Consensus in science
Nearly all (around 97%) of the scientific community accepts evolution as the dominant scientific theory of biological diversity. Atheists have used that data in an attempt to bolster their worldview and reject intelligent design inferences. You can beat consensus in science with one fact. But you can't convince an idiot about God's existence with a thousand facts. Much of the "settled science" that even geologists and other degree-holding "scientists" accept is really not established fact, it's only the most "widely accepted theory", and some actually ignore evidence that might support other and better inferences from available evidence, because that evidence indicates something other than the "consensus opinion" on a subject. Never mind that almost all the most groundbreaking and world-changing scientific and mathematical breakthroughs from Galileo to Newton, to Pasteur, to Pascal and Einstein, etc. were made by people who rejected conventional wisdom or went well beyond what "everybody knows".
Jorge R. Barrio corresponding in Consensus Science and the Peer Review 2009 Sep 11
I recently reviewed a lecture on science, politics, and consensus that Michael Crichton—a physician, producer, and writer—gave at the California Institute of Technology in Pasadena, CA, USA on January 17, 2003. I was struck by the timeliness of its content. I am quite certain that most of us have been—in one way or another—exposed to the concept (and consequences) of “consensus science.” In fact, scientific reviewers of journal articles or grant applications—typically in biomedical research—may use the term (e.g., “....it is the consensus in the field...”) often as a justification for shutting down ideas not associated with their beliefs. It begins with Stump's appeal to authority. This is a common evolutionary argument, but the fact that a majority of scientists accept an idea means very little. Certainly, expert opinion is an important factor and needs to be considered, but the reasons for that consensus also need to be understood. The history of science is full of examples of new ideas that accurately described and explained natural phenomena, yet were summarily rejected by experts. Scientists are people with a range of nonscientific, as well as scientific influences. Social, career, and funding influences are easy to underestimate. There can be tremendous pressures on a scientist that has little to do with the evidence at hand. This certainly is true in evolutionary circles, where the pressure to conform is intense. 12
1. Ernst Haeckel on the mechanical theory of life, and on spontaneous generation 1871
2. Lindsay Brownell: Embracing cellular complexity17 July 2014
3. Bharath Ramsundar: The Ferocious Complexity Of The Cell 2016
4. David Goodsell: Our Molecular Nature 1996
5. John D. Barrow: FITNESS OF THE COSMOS FOR LIFE, Biochemistry and Fine-Tuning
6. Kelly James Clark: Religion and the Sciences of Origins: Historical and Contemporary Discussions 2014
7. Paul Davies: The Goldilocks Enigma: Why Is the Universe Just Right for Life? April 29, 2008
8. Andreas Sommer: Materialism vs. Supernaturalism? “Scientific Naturalism” in Context July 19, 2018
9. Sean Carroll: The Big Picture: On the Origins of Life, Meaning, and the Universe Itself 10 may 2016
10. http://darwin-online.org.uk/content/frameset?itemID=F373&viewtype=image&pageseq=1
11. https://en.wikipedia.org/wiki/Intelligent_design
12. Jorge R. Barrio Consensus Science and the Peer Review 2009 Apr 28
13. https://www.amazon.com.br/Origins-Modern-Science-1300-1800/dp/0029050707
14. Prof. Dr. Ruth E. Blake: Special Issue 15 February 2020
15. Kepa Ruiz-Mirazo: [url=https://pubs.acs.org/doi/10.1021/cr2004844]Prebiotic Systems Chemistr
Last edited by Otangelo on Mon May 15, 2023 7:37 am; edited 111 times in total