https://reasonandscience.catsboard.com/t2131-the-mitochondrion
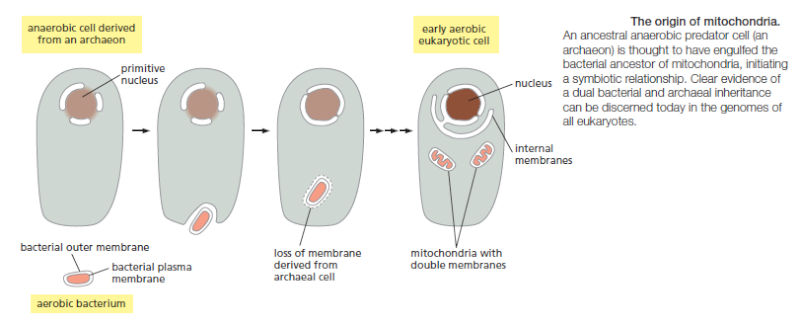
Endosymbiotic organelles, most notably mitochondria and chloroplasts, arguably represent the penultimate stage of cellular degradation. These organelles retain their own genomes, albeit with very few genes, their own internal translation systems and their own membranes, although substantially modified from the ancestral bacterial membranes.
Proteomic studies have revealed that mitochondria contain more than 1,000 different proteins, more than 99% of which are encoded by nuclear DNA. Thus, these proteins are synthesized as precursors on cytosolic ribosomes, specifically targeted to mitochondria and sorted into one of the four mitochondrial subcompartments. Various protein translocation machinery of the inner and outer mitochondrial membranes accomplish the complicated task of selective protein sorting into and across lipid bilayers. 2

James Villegas The mitochondria is an organelle (cellular organ) within a cell that takes organic sugar/ fuel molecules such as glucose (a product that results from digesting food) and converts said molecule(s) into an energy molecule known as ATP (Adenosine Triphosphate), which gives us the energy to do everyday physical tasks. In order to produce this molecule, a few basic steps must occur ( Glycolysis, Krebs Cycle/Citric Acid Cycle, and the Electron Transport Chain all in that order). The real money-maker that generates the majority of ATP is the last cycle, which is illustrated in the image of the electron transport chain (ETC) in the link above. This points to a Creator in that there are various protein complexes (such as ATP Synthase which is the ROTOR/ GEAR-LIKE structure in the end of the chain) and membranes IN THE ETC ALONE that are specifically designed to efficiently carry out a specific task. Often times, structures that are as efficient and complex as this are often existent due to the fact that they needed to be assembled. If the assemble of an intricate structure like this was left up to chance, the possibility of it being as efficient as it is now would be very low. This would be like asking a fully working engine to be assembled through random air currents blowing the pieces together, which although theoretically is not an impossibility, it is highly improbable. The same concept applies to this organic structure, although there is a possibility for a complex mechanism such as this to form independently isn't necessarily impossible, the probability of this doing so is HIGHLY UNLIKELY and thus needs to be intelligently designed if it is to work as efficiently as it does today.
To maintain their high degree of organization in a universe that is constantly drifting toward chaos, cells have a constant need for a plentiful supply of ATP. In eukaryotic cells, most of the ATP that powers life processes is produced by specialized, membrane-enclosed, energy-converting organelles. These are of two types. Mitochondria, which occur in virtually all cells of animals, plants, and fungi, burn food molecules to produce ATP by oxidative phosphorylation. Chloroplasts, which occur only in plants and green algae, harness solar energy to produce ATP by photosynthesis. In electron micrographs, the most striking features of both mitochondria and chloroplasts are their extensive internal membrane systems. These internal membranes contain sets of membrane protein complexes that work together to produce most of the cell’s ATP. In bacteria, simpler versions of essentially the same protein complexes produce ATP, but they are located in the cell’s plasma membrane

chemiosmotic coupling, signifying a link between the chemical bond-forming reactions that generate ATP (“chemi”) and membrane transport processes (“osmotic”). The chemiosmotic process occurs in two linked stages, both of which are performed by protein complexes in a membrane.
Stage 1: High-energy electrons (derived from the oxidation of food molecules, from pigments excited by sunlight, or from other sources ) are transferred along a series of electron-transport protein complexes that form an electron-transport chain embedded in a membrane. Each electron transfer releases a small amount of energy that is used to
pump protons (H+) and thereby generate a large electrochemical gradient across the membrane. Such an electrochemical gradient provides a way of storing energy, and it can be
harnessed to do useful work when ions flow back across the membrane.
Stage 2: The protons flow back down their electrochemical gradient through an elaborate membrane protein machine called ATP synthase, which catalyzes the production of ATP from ADP and inorganic phosphate (Pi). This ubiquitous enzyme works like a turbine in the membrane, driven by protons, to synthesize ATP . In this way, the energy derived
from food or sunlight in stage 1 is converted into the chemical energy of a phosphate bond in ATP.
Electrons move through protein complexes in biological systems via tightly bound metal ions or other carriers that take up and release electrons easily, or by special small molecules that pick electrons up at one location and deliver them to another. For mitochondria, the first of these electron carriers is NAD+, a water-soluble small molecule that takes up two electrons and one H+ derived from food molecules (fats and carbohydrates) to become NADH. NADH transfers these electrons from the sites where the food molecules are degraded to the inner mitochondrial membrane. There, the electrons from the energy-rich NADH are passed from one membrane protein complex to the next, passing to a lower-energy compound at each step, until they reach a final complex in which they combine with molecular oxygen (O2) to produce water. The energy released at each step as the electrons flow down this path from the energy-rich NADH to the low-energy water molecule drives H+ pumps in the inner mitochondrial membrane, utilizing three
different membrane protein complexes. Together, these complexes generate the proton-motive force harnessed by ATP synthase to produce the ATP that serves as the universal energy currency throughout the cell.
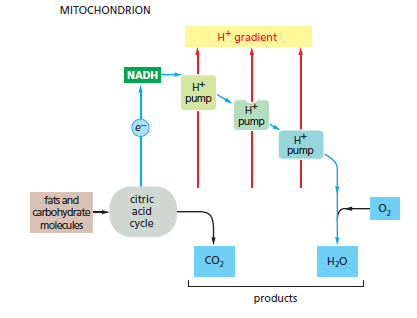
Electron-transport processes In the mitochondrion, fats and carbohydrates from food molecules are fed into the citric acid cycle and provide electrons to generate the energy-rich compound NADH from NAD+. These electrons then flow down an energy gradient as they pass from one complex to the next in the electron-transport chain, until they combine with molecular O2 in the last complex to produce water. The energy released at each stage is harnessed to pump H+ across the membrane.
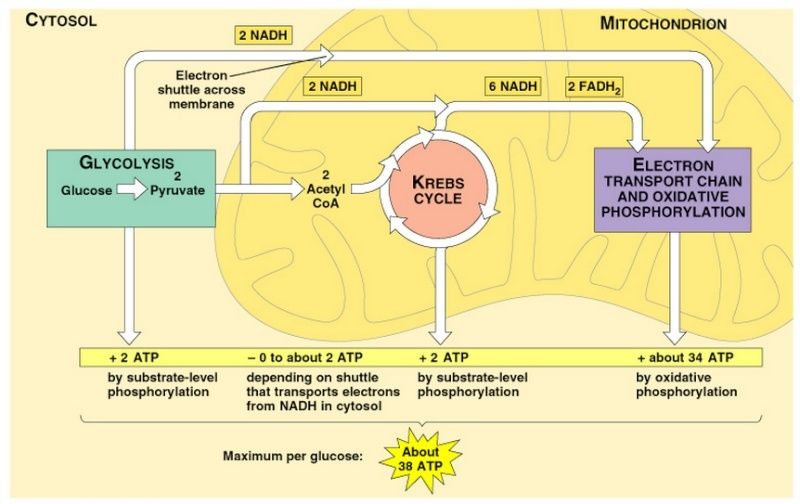
Mitochondria occupy up to 20% of the cytoplasmic volume of a eukaryotic cell. Although they are often depicted as short, bacterium-like bodies with a diameter of 0.5–1 μm, they are in fact remarkably dynamic and plastic, moving about the cell, constantly changing shape, dividing, and fusing. Mitochondria are often associated with the microtubular cytoskeleton, which determines their orientation and distribution in different cell types. Thus, in highly polarized cells such as neurons, mitochondria can move long distances (up to a meter or more in the extended axons of neurons), being propelled along the tracks of the microtubular cytoskeleton. In other cells, mitochondria remain fixed at points of high energy demand; for example, in skeletal or cardiac muscle cells, they pack between myofibrils, and in sperm cells they wrap tightly around the flagellum. Mitochondria also interact with other membrane systems in the cell, most notably the endoplasmic reticulum (ER). Contacts between mitochondria and ER define specialized domains thought to facilitate the exchange of lipids between the two membrane systems. These contacts also appear to induce mitochondrial fission, which is involved in the distribution and partitioning of mitochondria within cells. Mitochondria is a prerequisite for complex animals. Without mitochondria, present-day animal cells would have had to generate all of their ATP through anaerobic glycolysis. When Glycolysis converts glucose to pyruvate, it releases only a small fraction of the total free energy that is potentially available from glucose oxidation . In mitochondria, the metabolism of sugars is complete: pyruvate is imported into the mitochondrion and ultimately oxidized by O2 to CO2 and H2O, which allows 15 times more ATP to be made from a sugar than by glycolysis alone. This became possible only when enough molecular oxygen accumulated in the Earth’s atmosphere to allow organisms to take full advantage, via respiration, of the large amounts of energy potentially available from the oxidation of organic compounds. Mitochondria are large enough to be seen in the light microscope.
Where the inner membrane runs parallel to the outer membrane, between the cristae, it is known as the inner boundary membrane. The narrow (20–30 nm) gap between the inner boundary membrane and the outer membrane is known as the intermembrane space. The cristae are about 20 nm-wide membrane discs or tubules that protrude deeply into the matrix and enclose the crista space. The crista membrane is continuous with the inner boundary membrane, and where their membranes join, the membrane forms narrow membrane tubes or slits, known as crista junctions. Like the bacterial outer membrane, the outer mitochondrial membrane is freely permeable to ions and to small molecules as large as 5000 daltons. This is because it contains many porin molecules, a special class of β-barrel-type membrane protein that creates aqueous pores across the membrane. As a consequence, the intermembrane space between the outer and inner membrane has the same pH and ionic composition as the cytoplasm, and there is no electrochemical gradient across the outer membrane.
The Mitochondrion Has an Outer Membrane and an Inner Membrane
Mitochondria have an outer and an inner membrane. The two membranes have distinct functions and properties and delineate separate compartments within the organelle. The inner membrane, which surrounds the internal mitochondrial matrix compartment is highly folded to form invaginations known as cristae (the singular is crista), which contain in their membranes the proteins of the electron-transport chain.

Picture above: Structure of a mitochondrion. (A) The outer membrane envelops the inner boundary membrane. The inner membrane is highly folded into tubular or lamellar cristae, which crisscross the matrix. The dense matrix, which contains most of the mitochondrial protein, appears dark in the electron microscope, whereas the intermembrane space and the crista space appear light due to their lower protein content. The inner boundary membrane follows the outer membrane closely at a distance of ≈20 nm. The inner membrane turns sharply at the cristae junctions, where the cristae join the inner boundary membrane. (C) Schematic drawing of a mitochondrion showing the outer membrane (gray), and the inner membrane (yellow). Note that the inner membrane is compartmentalized into the inner boundary membrane and the crista membrane. There are three distinct spaces: the inner membrane space, the crista space, and the matrix.
The matrix contains the genetic system of the mitochondrion, including the mitochondrial DNA and the ribosomes. The mitochondrial DNA is organized into compact bodies—the nucleoids—by special scaffolding proteins that also function as transcription regulatory proteins. The large number of enzymes required for the maintenance of the mitochondrial
genetic system, as well as for many other essential reactions to be outlined next, accounts for the very high protein concentration in the matrix; at more than 500 mg/mL, this concentration is close to that in a protein crystal.
Mitochondria Have Many Essential Roles in Cellular Metabolism
Mitochondria not only generate most of the cell’s ATP; they also provide many other essential resources for biosynthesis and cell growth. Before describing in detail the remarkable machinery of the respiratory chain, we diverge briefly to touch on some of these important roles. Mitochondria are critical for buffering the redox potential in the cytosol. Cellsneed a constant supply of the electron acceptor NAD+ for the central reaction in glycolysis that converts glyceraldehyde 3-phosphate to 1,3-bisphosphoglycerate. This NAD+ is converted to NADH in the process, and the NAD+ needs to be regenerated by transferring the high-energy NADH electrons somewhere. The NADH electrons will eventually be used to help drive oxidative phosphorylation inside the mitochondrion. But the inner mitochondrial membrane is impermeable to NADH. The electrons are therefore passed from the NADH to smaller molecules in the cytosol that can move through the inner mitochondrial membrane. Once in the matrix, these smaller molecules transfer their electrons to NAD+ to form mitochondrial NADH, after which they are returned to the cytosol for recharging—creating a so-called shuttle system for the NADH electrons. In addition to ATP, biosynthesis in the cytosol requires both a constant supply of reducing power in the form of NADPH and small carbon-rich molecules to serve as building blocks. Descriptions of biosynthesis often state that the needed carbon skeletons come directly from the breakdown of sugars, whereas the NADPH is produced in the cytosol by a side pathway for the breakdown of sugars (the pentose phosphate pathway, an alternative to glycolysis). But under conditions where nutrients abound and plenty of ATP is available, mitochondria help to generate both the reducing power and the carbon- rich building blocks needed for cell growth. For this purpose, excess citrate produced in the mitochondrial matrix by the citric acid cycle is transported down its electrochemical gradient to the cytosol, where it is metabolized to produce essential components of the cell. Thus, for example, as part of a cell’s response to growth signals, large amounts of acetyl CoA are produced in the cytosol from citrate exported from mitochondria, accelerating the production of the fatty acids
and sterols that build new membranes .
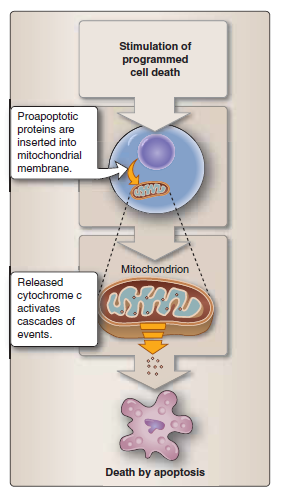
The urea cycle is a central metabolic pathway in mammals that converts the ammonia (NH4 +) produced by the breakdown of nitrogen-containing compounds (such as amino acids) to the urea excreted in urine. Two critical steps of the urea cycle are carried out in the mitochondria of liver cells, while the remaining steps occur in the cytosol. Mitochondria also play an essential part in the metabolic adaptation of cells to different nutritional conditions. For example, under conditions of starvation, proteins in our bodies are broken down to amino acids, and the amino acids are imported into mitochondria and oxidized to produce NADH for ATP production. The biosynthesis of heme groups—which, as we shall see in the next section, play a central part in electron transfer—is another critical process that is shared between the mitochondrion and the cytoplasm. Iron–sulfur clusters, which are essential not only for electron transfer in the respiratory chain, but also for the maintenance and stability of the nuclear genome, are produced in mitochondria (and chloroplasts). Nuclear genome instability, a hallmark of cancer, can sometimes be linked to the decreased function of cellular proteins that contain iron–sulfur clusters. Mitochondria also have a central role in membrane biosynthesis. Cardiolipin is a two-headed phospholipid that is confined to the inner mitochondrial membrane, where it is also produced. But mitochondria are also a major source of phospholipids for the biogenesis of other cell membranes. Phosphatidylethanolamine, phosphatidylglycerol, and phosphatidic acid are synthesized in the mitochondrion, while phosphatidylinositol, phosphatidylcholine, and phosphatidylserine are primarily synthesized in the endoplasmic reticulum (ER). Most of the cell’s membranes are assembled in the ER. The exchange of lipids between the ER and mitochondria is thought to occur at special sites of close contact by an as-yet unknown mechanism. Finally, mitochondria are important calcium buffers, taking up calcium from the ER and sarcoplasmic reticulum at special membrane junctions. Cellular calcium levels control muscle contraction and alterations are implicated in neurodegeneration and apoptosis. Clearly, cells and organisms depend on mitochondria in many different ways. We now return to the central function of the mitochondrion in respiratory ATP generation.
The Inner Membrane Cristae Contain the Machinery for Electron Transport and ATP Synthesis
Unlike the outer mitochondrial membrane, the inner mitochondrial membrane is a diffusion barrier to ions and small molecules, just like the bacterial inner membrane. However, selected ions, most notably protons and phosphate, as well as essential metabolites such as ATP and ADP, can pass through it by means of special transport proteins. The inner mitochondrial membrane is highly differentiated into functionally distinct regions with different protein compositions. The lateral segregation of membrane regions with different protein and lipid compositions is a key feature of cells. In the inner mitochondrial membrane, the boundary membrane region is thought to contain the machinery for protein import, new membrane insertion, and assembly of the respiratory-chain complexes. The membranes of the cristae, which are continuous with the boundary membrane, contain the ATP synthase enzyme that produces most of the cell’s ATP; they also contain the large protein complexes of the respiratory chain—the name given to the mitochondrion’s electron-transport chain. At the cristae junctions, where the membranes of the cristae join the boundary membrane, special protein complexes provide a diffusion barrier that segregates the membrane proteins in the two regions of the inner membrane; these complexes are also thought to anchor the cristae to the outer membrane, thus maintaining the highly folded topology of the inner membrane. Cristae membranes have one of the highest protein densities of all biological membranes, with a lipid content of 25% and a protein content of 75% by weight. The folding of the inner membrane into cristae greatly increases the membrane area available for oxidative phosphorylation. In highly active cardiac muscle cells, for example, the total area of cristae membranes can be up to 20 times larger than the area of the cell’s plasma membrane. In total, the surface area of cristae membranes in each human body adds up to roughly the size of a football field.
The Citric Acid Cycle in the Matrix Produces NADH
Together with the cristae that project into it, the matrix is the major working part of the mitochondrion. Mitochondria can use both pyruvate and fatty acids as fuel. Pyruvate is derived from glucose and other sugars, whereas fatty acids are derived from fats. Both of these fuel molecules are transported across the inner mitochondrial membrane by specialized transport proteins, and they are then converted to the crucial metabolic intermediate acetyl-CoA by enzymes located in the mitochondrial matrix.
The acetyl groups in acetyl-CoA are oxidized in the matrix via the citric acid cycle, also called the Krebs cycle. The oxidation of these carbon atoms in acetyl CoA produces CO2, which diffuses out of the mitochondrion to be released to the environment as a waste product. More importantly, the citric acid cycle saves a great deal of the bond energy released by this oxidation in the form of electrons carried by NADH. This NADH transfers its electrons from the matrix to the electron-transport chain in the inner mitochondrial
membrane, where—through the chemiosmotic coupling process the energy that was carried by NADH electrons is converted into phosphate-bond energy in ATP.
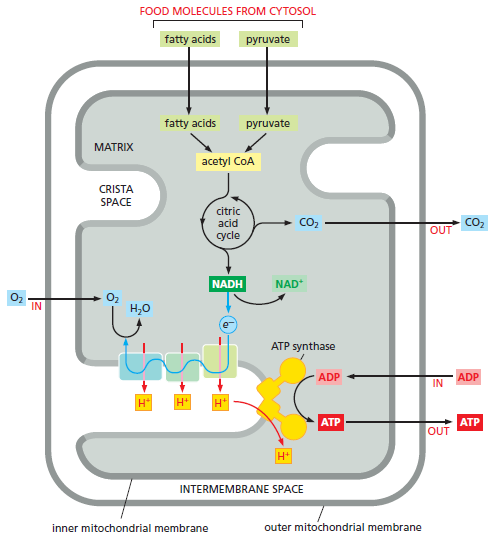
Picture above: A summary of the energy converting metabolism in mitochondria. Pyruvate and fatty acids enter the mitochondrion (top of the figure) and are broken down to acetyl CoA. The acetyl-CoA is metabolized by the citric acid cycle, which reduces NAD+ to NADH, which then passes its high-energy electrons to the first complex in the electron-transport chain. In the process of oxidative phosphorylation, these electrons pass along the electron transport chain in the inner membrane cristae to oxygen (O2). This electron transport generates a proton gradient, which drives the production of ATP by the ATP synthase. Electrons from the oxidation of succinate, a reaction intermediate in the citric acid cycle, take a separate path to enter this electron-transport chain The membranes that comprise the mitochondrial inner membrane—the inner boundary membrane and the crista membrane—contain different mixtures of proteins and they are therefore shaded differently in this diagram.
A Chemiosmotic Process Couples Oxidation Energy to ATP Production
Although the citric acid cycle that takes place in the mitochondrial matrix is considered to be part of aerobic metabolism, it does not itself use oxygen. Only the final step of oxidative metabolism consumes molecular oxygen (O2) directly. Nearly all the energy available from metabolizing carbohydrates, fats, and other foodstuffs in earlier stages is saved in the form of energy-rich compounds that feed electrons into the respiratory chain in the inner mitochondrial membrane. These electrons, most of which are carried by NADH, finally combine with O2 at the end of the respiratory chain to form water. The energy released during the complex series of electron transfers from NADH to O2 is harnessed in the inner membrane to generate an electrochemical gradient that drives the conversion of ADP + Pi to ATP. For this reason, the term oxidative phosphorylation is
used to describe this final series of reactions.

The total amount of energy released by biological oxidation in the respiratory chain is equivalent to that released by the explosive combustion of hydrogen when it combines with oxygen in a single step to form water. But the combustion of hydrogen in a single-step chemical reaction, which has a strongly negative ΔG, releases this large amount of energy unproductively as heat. In the respiratory chain, the same energetically favorable reaction H2 + ½ O2 → H2O is divided into small steps.

This stepwise process allows the cell to store nearly half of the total energy that is released in a useful form. At each step, the electrons, which can be thought of as having been removed from a hydrogen molecule to produce two protons, pass through a series of electron carriers in the inner mitochondrial membrane. At each of three distinct steps along the way (marked by the three electron-transport complexes of the respiratory chain), much of the energy is utilized for pumping protons across the membrane. At the end of the
electron-transport chain, the electrons and protons recombine with molecular oxygen into water. Water is a very low-energy molecule and is thus very stable; it can serve as anelectron donor only when a large amount of energy from an external source is spent on splitting it into protons, electrons, and molecular oxygen. This is exactly what happens in oxygenic photosynthesis, where the external energy source is the sun.
The Energy Derived from Oxidation Is Stored as an Electrochemical Gradient
In mitochondria, the process of electron transport begins when two electrons and a proton are removed from NADH (to regenerate NAD+). These electrons are passed to the first of about 20 different electron carriers in the respiratory chain. The electrons start at a large negative redox potential —that is, at a high energy level—which gradually drops as they pass along the chain. The proteins involved are grouped into three large respiratory enzyme complexes, each composed of protein subunits that sit in the inner mitochondrial membrane. Each complex in the chain has a higher affinity for electrons than its predecessor, and electrons pass sequentially from one complex to the next until they are finally
transferred to molecular oxygen, which has the highest electron affinity of all. The net result is the pumping of H+ out of the matrix across the inner membrane, driven by the energetically favorable flow of electrons. This transmembrane movement of H+ has two major consequences:
1. It generates a pH gradient across the inner mitochondrial membrane, with a high pH in the matrix (close to) and a lower pH in the intermembrane space. Since ions and small molecules equilibrate freely across the outer mitochondrial membrane, the pH in the intermembrane space is the same as in the cytosol (generally around pH 7.4).
2. It generates a voltage gradient across the inner mitochondrial membrane, creating a membrane potential with the matrix side negative and the crista space side positive. The pH gradient (ΔpH) reinforces the effect of the membrane potential (ΔV ), because the latter acts to attract any positive ion into the matrix and to push any negative ion out. Together, ΔpH and ΔV make up the electrochemical gradient, which is measured in units of millivolts (mV). This gradient exerts a protonmotive force, which tends to drive H+ back into the matrix (Figure below )

1) http://www.sciencedirect.com/science/article/pii/S0167488912001449
2) https://www.biochemie.uni-freiburg.de/ag/vanderLaan/research?set_language=en
Last edited by Otangelo on Tue Jul 26, 2022 7:43 am; edited 26 times in total