Protein Complexes Shape Chromatin's 3D Structure
The 3D organization of chromatin is sculpted by an extensive array of protein complexes that craft loops, vital for regulating transcription from DNA to mRNA. These protein assemblies not only facilitate the formation of loops but also anchor the chromatin within specific nuclear locales. Among these, three principal complexes—insulator protein CTCF, cohesion, and Mediator—play pivotal roles in molding the 3D chromatin architecture.
The insulator protein CTCF is instrumental in bridging long-distance chromatin interactions near Topologically Associating Domains (TADs), essentially laying the groundwork for TAD structure. While some CTCF molecules function as insulators, keeping distant chromatin regions separate, others do not. CTCF's multifaceted roles include forming DNA insulators to prevent enhancer-promoter interactions, anchoring chromatin loops to the nuclear lamina for spatial organization, demarcating active from inactive chromatin regions, and thereby influencing the genetic networks active in a cell.
Cohesion, another crucial complex, shapes chromatin by forming loops that vary by cell type and are associated with enhancers. It plays a continuous role, not only by encircling sister chromatids post-replication to maintain their cohesion but also by facilitating chromatin connections during cell division. Cohesins are pivotal for chromosome spindle attachment, DNA recombination—a cornerstone of evolutionary change and DNA repair—and transcription regulation.
These protein complexes, through their diverse functions and interactions, orchestrate the complex 3D conformation of chromatin, impacting gene expression and cellular function. Humans and worms share a surprising similarity in their genetic makeup, both possessing around 21,000 genes. Despite this numerical equivalence, the vast diversity in cellular functions across different organisms and even within the various cell types of a single organism is not dictated merely by the genes themselves. Instead, the key lies in how these genes are utilized and expressed differently across cell types, leading to the production of distinct proteins and cellular structures.
Epigenetics shed light on the mechanisms that operate beyond the straightforward genetic code-to-protein translation, revealing a complex landscape of gene regulation. Recent advances have uncovered that the three-dimensional organization of the cell nucleus plays a crucial role in gene function. The dynamic 3D shapes of chromatin, the complex of DNA and protein found in the nucleus, along with specific DNA packaging and localization, are essential for the regulation of gene activity. DNA regulation occurs at multiple levels, one of which involves the editing of DNA to produce various messenger RNAs from the same gene segment, by alternative RNA splicing. The ENCODE project has further blurred the traditional boundaries of what constitutes a gene, showing that segments from different DNA regions can be combined to produce a single protein. This discovery challenges the conventional definition of a gene. The structural basis for DNA organization involves histones, around which DNA is wound like a thread on a spool. These histones, along with the DNA, are marked by a variety of chemical modifications far more diverse than the commonly known acetyl and methyl groups. Over 30 different types of modifications, including multiple forms of methylation and processes like sumoylation and ubiquitination, contribute to the regulation of genetic activity. These modifications, along with the formation of specific 3D structures facilitated by large protein complexes, influence the activation of genetic networks in response to different cellular needs.
Adding to the complexity, numerous non-coding RNAs, which do not translate into proteins, play a pivotal role in gene regulation. Initially considered "junk" DNA, these non-coding regions are now recognized for producing RNAs that regulate gene expression in various ways. The regulatory capacity of these RNAs suggests that the non-protein-coding portion of the genome is far more significant than previously thought, potentially outnumbering protein-coding genes by a substantial margin. The spatial organization of chromosomes within the nucleus also impacts gene function. The remarkable feat of fitting approximately 2 meters of DNA into the microscopic nucleus is achieved through intricate folding and compaction strategies, allowing for rapid gene expression changes as required, for instance, in neurons responding to stimuli. The relationship between chromosomal architecture and gene activity is multifaceted, with chromatin loops, nuclear positioning, and interactions with the nuclear lamina all playing roles in gene regulation.
Histones, the structural backbone of chromatin, come in various families and form nucleosomes, which are fundamental units of DNA packaging resembling a yoyo with DNA wrapped around it. These nucleosomes link together to form complex structures that not only protect DNA but also regulate gene accessibility and activity, highlighting the intricate interplay between chromatin structure and gene expression. Histones H3 and H4 possess extensive tails that serve as sites for epigenetic modifications through various processes such as acetylation, methylation, phosphorylation, ubiquitination, SUMOylation, citrullination, and ribosylation. Even the core parts of these histones can undergo diverse modifications, each altering gene function in their vicinity and contributing to a complex "histone code." These modifications are crucial for DNA repair, gene regulation, chromosome structuring, and are often passed down to subsequent generations. Recent research suggests that methylation patterns established during development are pivotal in shaping the female brain. The term "chromatin" describes the complex assembly of nucleosomes. Initially observed under a microscope, chromatin was categorized into two types: heterochromatin and euchromatin, though further research has revealed more nuanced subtypes.
Heterochromatin, which is densely packed and generally inactive, is found at the nucleus's periphery. It encompasses at least five distinct states with varying epigenetic markers and includes regions like telomeres and centromeres. Constitutive heterochromatin contains repetitive elements forming structures like centromeres, while facultative heterochromatin consists of silenced genes that can potentially be reactivated. Euchromatin, in contrast, is the site of active gene expression, enriched with genes, RNAs, and proteins, and is positioned closer to the nucleus's center. The nucleus itself harbors a complex architecture that is intricately linked to chromatin organization. The nucleolus, located near the center, is tasked with ribosome synthesis and assembly. The nuclear lamina, comprising intermediate filament proteins known as lamins, forms complex structures that organize chromatin and influence replication and cell division by anchoring specific chromatin regions. Topologically Associated Domains (TADs) represent higher-level DNA structures, akin to protein folding, where chromatin folds into regions forming TADs and larger structures, including loops, which contribute to chromosome architecture. These structures vary within individual cells, unlike the more fixed structures of most proteins. Chromatin loops facilitate gene regulation, with stem cells displaying less defined chromosomal structures that become more structured upon differentiation, thus limiting gene accessibility and defining cell identity. The density of nucleosome packaging, or "clutches," varies with cell differentiation, with stem cells having more loosely packed nucleosomes.
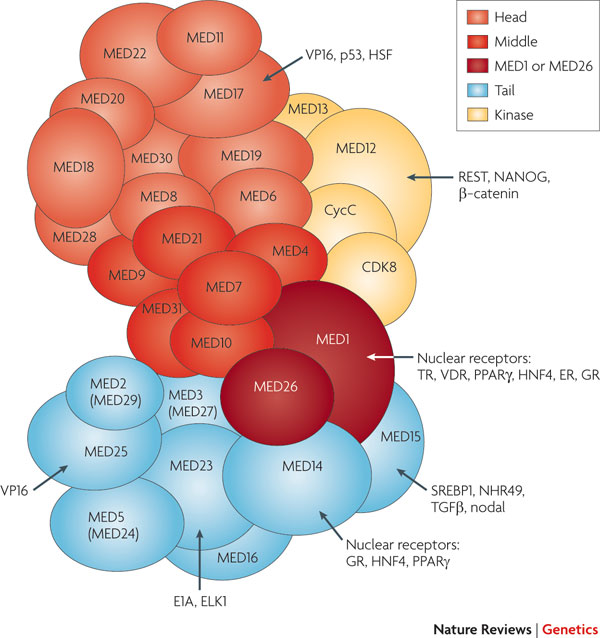
Protein complexes such as CTCF, cohesin, and Mediator play pivotal roles in shaping 3D chromatin structures. CTCF organizes chromatin interactions and establishes boundaries between active and inactive chromatin regions. Cohesin complexes form loops that vary across cell types and are essential for chromosome cohesion during cell division. Mediator, a large multiprotein complex, is crucial for activating gene networks and facilitating transcription by connecting enhancers and promoters. TAD structures significantly impact genetic functions, with their organization reflecting regions of active and inactive DNA. The dynamic nature of TADs, especially in facultative chromatin, allows for shifts during development, illustrating the interplay between the histone code and 3D chromatin architecture in regulating gene expression.
Secondary Structures
Chromatin structures differentiate to form distinct regions known as Topologically Associating Domains (TADs), driven by various factors that maintain their localization. The chromatin architecture varies significantly between genes essential for ongoing cellular functions and those active only during fetal development, the latter being relegated to inactive chromatin regions post-development. Transcription is a key player in shaping the three-dimensional TAD structures, with RNA polymerase playing a role in elongating DNA and altering chromatin configurations. Active chromatin regions feature interactions between enhancers and promoters, and active promoters often engage with other promoters, forming loop structures around start and stop sites. Polycomb sites contribute to another layer of structural organization by forming clusters within a TAD that highlight specific interactions, aiding in the chromatin's folding. Certain DNA sequences play a role in these three-dimensional structures, although not all sequences contribute equally. Various diseases are linked to the loss of specific DNA sequences, leading to alterations in the 3D chromatin structure, which can influence disease pathology. During cell division, there is precise regulation of 3D chromatin structures. TADs are present during interphase but not mitosis, indicating their evolutionary significance, as evidenced by their conservation across species for over 40 million years, highlighting their potential role in evolutionary processes.
Tertiary Structure
TADs play a crucial role in shaping large-scale chromosome structures, with some TADs engaging in long-range interactions with other chromosome regions. These structural variants are observed even within similar cell populations of an organism. Chromosomes exhibit dynamic movements within the nuclear space, constrained to specific areas. Certain chromosomes associate with nucleoli or the nuclear periphery, and coordinated long-range movements of multiple chromosomes are observed. Stable long-range interactions serve specific functions, such as the activation or repression of transcription factors, exemplified by the long-range interactions between three genes associated with the cytokine TNFα. The 3D chromatin structure fosters numerous associations, including DNA loop bonding, close relationships between co-regulated genes, and the spatial organization of active and repressed chromatin regions. TAD structures ensure the integrity of genetic networks and enhance the efficacy of regulatory factors in protein production.
Vast Complexity of Chromatin 3D Shapes
The complexity of genetic regulation is rapidly expanding, with mechanisms such as alternative RNA splicing, RNA silencing, and the involvement of various non-coding RNAs. Adding to this complexity are the higher-order 3D structures of chromatin and chromosomes, which are crucial for cell-specific gene network functions. Chromatin exhibits a highly organized yet dynamic 3D conformation, maintaining essential gene-regulatory interactions. These structural dynamics enable diverse genetic networks to function within the intricately organized yet compartmentalized nuclear space. Predicting chromatin's 3D structures remains a challenge, akin to predicting protein structures from amino acid sequences, with numerous protein complexes contributing to the chromatin architecture. This sophisticated organization and regulation of chromatin raise questions about the underlying directives, which cannot be solely attributed to DNA. The coordinated regulation of DNA and activation of gene networks, especially in the context of neuronal activity and thought processes, suggests a level of complexity beyond the DNA sequence, pointing to a multifaceted regulatory landscape in cellular and genetic function.